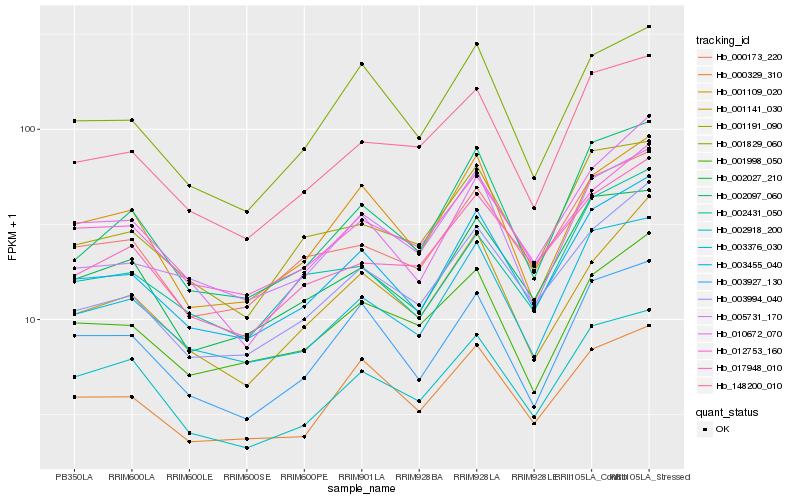
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003455_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_17270 [Jatropha curcas] |
| 2 |
Hb_003376_030 |
0.0525790818 |
- |
- |
PREDICTED: ribonuclease P protein subunit p29 isoform X2 [Jatropha curcas] |
| 3 |
Hb_003994_040 |
0.0552303672 |
- |
- |
- |
| 4 |
Hb_000173_220 |
0.0584679032 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 5 |
Hb_012753_160 |
0.0643267643 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 6 |
Hb_002027_210 |
0.0655800747 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 7 |
Hb_002097_060 |
0.0662502824 |
- |
- |
PREDICTED: 50S ribosomal protein L9, chloroplastic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000329_310 |
0.0666187443 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_005731_170 |
0.068887537 |
- |
- |
hypothetical protein PRUPE_ppa011750mg [Prunus persica] |
| 10 |
Hb_003927_130 |
0.0703230322 |
- |
- |
PREDICTED: nudix hydrolase 9 [Phoenix dactylifera] |
| 11 |
Hb_010672_070 |
0.0706983126 |
- |
- |
hypothetical protein ZEAMMB73_607937, partial [Zea mays] |
| 12 |
Hb_001191_090 |
0.0708463627 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 13 |
Hb_001829_060 |
0.071298086 |
- |
- |
Eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 14 |
Hb_148200_010 |
0.0739113127 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 15 |
Hb_001141_030 |
0.0741612009 |
- |
- |
hypothetical protein PRUPE_ppa024854mg [Prunus persica] |
| 16 |
Hb_001109_020 |
0.0751372561 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_017948_010 |
0.0753214271 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 18 |
Hb_002918_200 |
0.0764875648 |
- |
- |
- |
| 19 |
Hb_002431_050 |
0.0770649678 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 [Jatropha curcas] |
| 20 |
Hb_001998_050 |
0.0776936464 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |