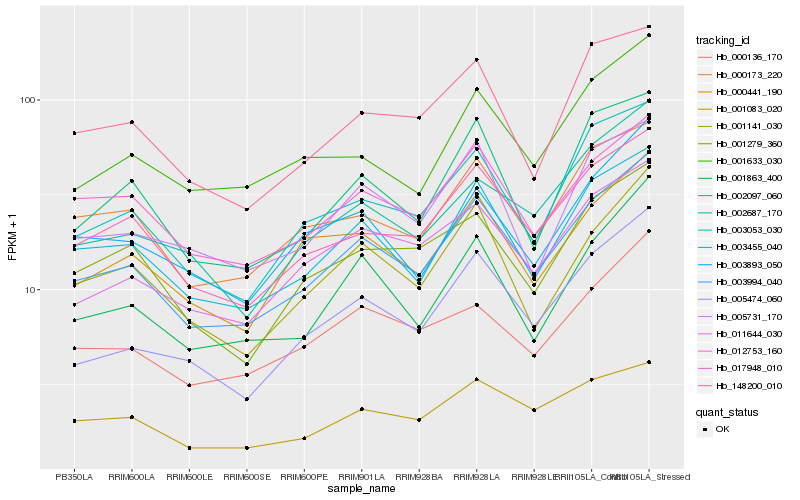
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003994_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_003455_040 |
0.0552303672 |
- |
- |
hypothetical protein JCGZ_17270 [Jatropha curcas] |
| 3 |
Hb_017948_010 |
0.0592331435 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 4 |
Hb_003053_030 |
0.0676294067 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 5 |
Hb_005474_060 |
0.069995776 |
- |
- |
hypothetical protein PHAVU_011G019700g [Phaseolus vulgaris] |
| 6 |
Hb_000173_220 |
0.0701178546 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 7 |
Hb_011644_030 |
0.0723063122 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Pyrus x bretschneideri] |
| 8 |
Hb_001863_400 |
0.0758414508 |
- |
- |
hypothetical protein EUGRSUZ_F03302 [Eucalyptus grandis] |
| 9 |
Hb_001141_030 |
0.0768093893 |
- |
- |
hypothetical protein PRUPE_ppa024854mg [Prunus persica] |
| 10 |
Hb_012753_160 |
0.0776701652 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 11 |
Hb_000441_190 |
0.0777528356 |
- |
- |
PREDICTED: GDP-L-galactose phosphorylase 1-like isoform X2 [Vitis vinifera] |
| 12 |
Hb_000136_170 |
0.0784235943 |
- |
- |
PREDICTED: uncharacterized protein LOC105645501 [Jatropha curcas] |
| 13 |
Hb_002097_060 |
0.0795685746 |
- |
- |
PREDICTED: 50S ribosomal protein L9, chloroplastic-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002687_170 |
0.0815496594 |
- |
- |
PREDICTED: G patch domain and ankyrin repeat-containing protein 1 homolog [Jatropha curcas] |
| 15 |
Hb_148200_010 |
0.0818258955 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 16 |
Hb_001279_360 |
0.0824262538 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_001083_020 |
0.0825248712 |
- |
- |
- |
| 18 |
Hb_003893_050 |
0.0830734954 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001633_030 |
0.083236092 |
- |
- |
- |
| 20 |
Hb_005731_170 |
0.083305475 |
- |
- |
hypothetical protein PRUPE_ppa011750mg [Prunus persica] |