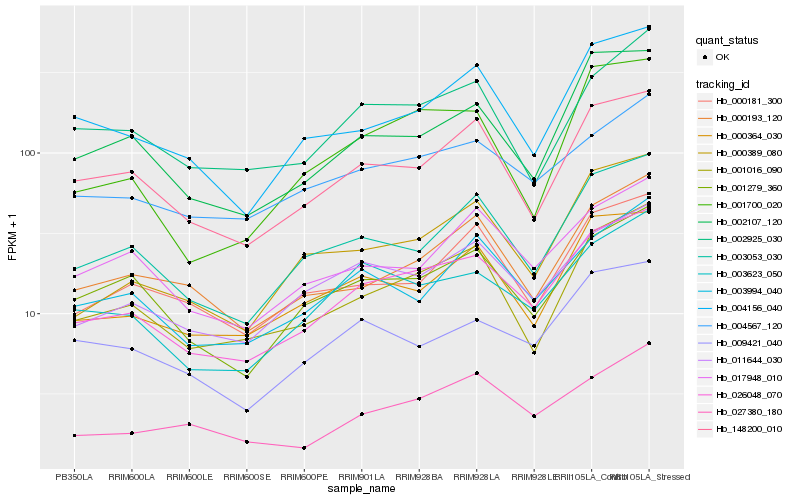
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026048_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 2 |
Hb_011644_030 |
0.0752709194 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Pyrus x bretschneideri] |
| 3 |
Hb_003623_050 |
0.0776558518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001016_090 |
0.0785493327 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 1 [Populus euphratica] |
| 5 |
Hb_003053_030 |
0.0818295528 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 6 |
Hb_148200_010 |
0.0846627532 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 7 |
Hb_001279_360 |
0.0860068859 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000364_030 |
0.087009512 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_004567_120 |
0.0890041393 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 10 |
Hb_002107_120 |
0.0896129753 |
- |
- |
PREDICTED: transcription elongation factor 1 homolog [Jatropha curcas] |
| 11 |
Hb_004156_040 |
0.0917223503 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 12 |
Hb_003994_040 |
0.0923904275 |
- |
- |
- |
| 13 |
Hb_000389_080 |
0.0930469491 |
- |
- |
PREDICTED: uncharacterized protein At4g26485-like [Jatropha curcas] |
| 14 |
Hb_000193_120 |
0.0970974691 |
- |
- |
PREDICTED: protein preY, mitochondrial [Jatropha curcas] |
| 15 |
Hb_027380_180 |
0.0976507655 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 16 |
Hb_009421_040 |
0.0977244733 |
- |
- |
PREDICTED: eukaryotic initiation factor 4A [Jatropha curcas] |
| 17 |
Hb_000181_300 |
0.0982457676 |
- |
- |
PREDICTED: uncharacterized protein LOC105650445 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002925_030 |
0.0984147609 |
- |
- |
40S ribosomal protein S16B [Hevea brasiliensis] |
| 19 |
Hb_001700_020 |
0.0985628136 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 20 |
Hb_017948_010 |
0.0985778972 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |