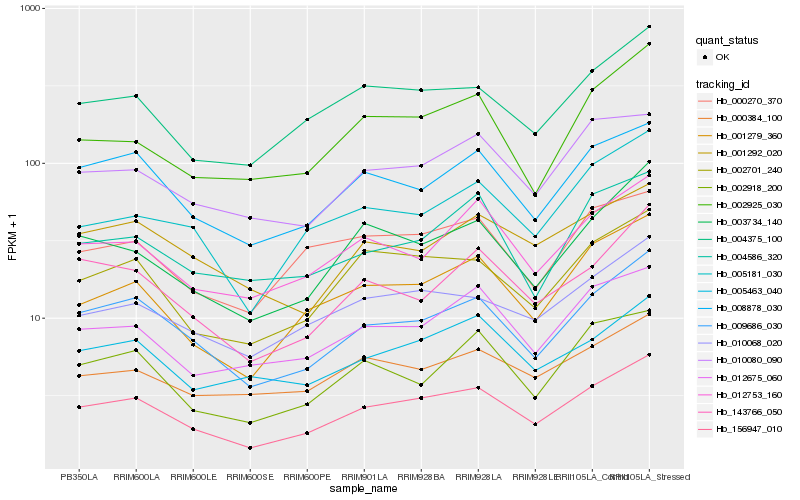
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_156947_010 |
0.0 |
- |
- |
kinase-like protein pac.x.7.4, partial [Platanus x acerifolia] |
| 2 |
Hb_009686_030 |
0.0530834039 |
- |
- |
- |
| 3 |
Hb_002701_240 |
0.0699078113 |
- |
- |
PREDICTED: putative DNA helicase INO80 [Jatropha curcas] |
| 4 |
Hb_004375_100 |
0.0722741075 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1 [Jatropha curcas] |
| 5 |
Hb_001279_360 |
0.0733425395 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_010068_020 |
0.075923105 |
- |
- |
PREDICTED: uncharacterized protein LOC105632966 [Jatropha curcas] |
| 7 |
Hb_008878_030 |
0.0767056448 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Musa acuminata subsp. malaccensis] |
| 8 |
Hb_012753_160 |
0.0819542234 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 9 |
Hb_012675_060 |
0.0826464581 |
- |
- |
hypothetical protein PRUPE_ppa007715mg [Prunus persica] |
| 10 |
Hb_003734_140 |
0.0834463742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010080_090 |
0.0867605059 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2 [Jatropha curcas] |
| 12 |
Hb_005463_040 |
0.0872149688 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 13 |
Hb_143766_050 |
0.0900126139 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-5-like [Populus euphratica] |
| 14 |
Hb_005181_030 |
0.0900482089 |
- |
- |
PREDICTED: huntingtin-interacting protein K [Jatropha curcas] |
| 15 |
Hb_000384_100 |
0.0915778326 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 16 |
Hb_000270_370 |
0.0934859573 |
- |
- |
Uncharacterized protein TCM_011954 [Theobroma cacao] |
| 17 |
Hb_002925_030 |
0.095477348 |
- |
- |
40S ribosomal protein S16B [Hevea brasiliensis] |
| 18 |
Hb_001292_020 |
0.0955137052 |
- |
- |
PREDICTED: keratin, type II cytoskeletal 5 isoform X1 [Sesamum indicum] |
| 19 |
Hb_004586_320 |
0.0956118791 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 20 |
Hb_002918_200 |
0.0967386079 |
- |
- |
- |