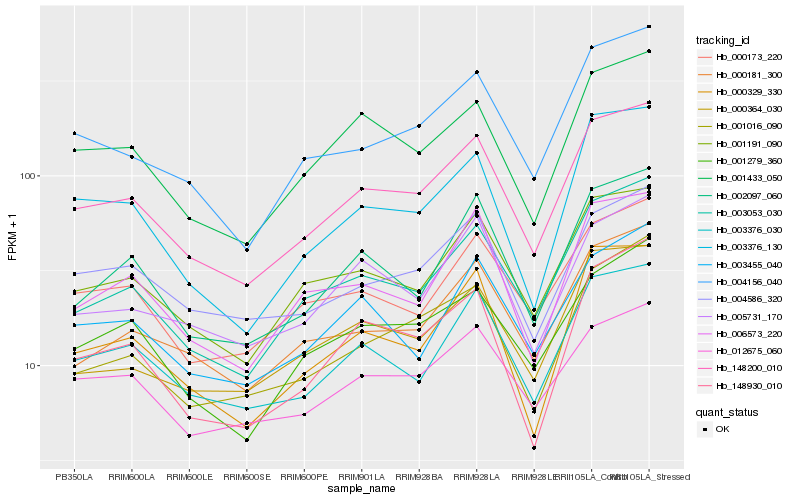
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148200_010 |
0.0 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 2 |
Hb_001191_090 |
0.05415546 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 3 |
Hb_003376_030 |
0.0572426372 |
- |
- |
PREDICTED: ribonuclease P protein subunit p29 isoform X2 [Jatropha curcas] |
| 4 |
Hb_003053_030 |
0.0574876352 |
- |
- |
PREDICTED: transport and Golgi organization 2 homolog [Jatropha curcas] |
| 5 |
Hb_001433_050 |
0.0604743151 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 6 |
Hb_004156_040 |
0.0645636672 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 7 |
Hb_001279_360 |
0.0647826745 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_002097_060 |
0.0683307667 |
- |
- |
PREDICTED: 50S ribosomal protein L9, chloroplastic-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_012675_060 |
0.0688519199 |
- |
- |
hypothetical protein PRUPE_ppa007715mg [Prunus persica] |
| 10 |
Hb_000329_330 |
0.0688519223 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 11 |
Hb_004586_320 |
0.0695395918 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 12 |
Hb_000181_300 |
0.0699034469 |
- |
- |
PREDICTED: uncharacterized protein LOC105650445 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000173_220 |
0.0715062926 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 14 |
Hb_006573_220 |
0.0726703179 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 15 |
Hb_005731_170 |
0.0728695691 |
- |
- |
hypothetical protein PRUPE_ppa011750mg [Prunus persica] |
| 16 |
Hb_148930_010 |
0.073120801 |
- |
- |
PREDICTED: uncharacterized protein LOC105648400 [Jatropha curcas] |
| 17 |
Hb_001016_090 |
0.0733876666 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 1 [Populus euphratica] |
| 18 |
Hb_003455_040 |
0.0739113127 |
- |
- |
hypothetical protein JCGZ_17270 [Jatropha curcas] |
| 19 |
Hb_003376_130 |
0.073993651 |
transcription factor |
TF Family: S1Fa-like |
DNA-binding protein S1FA, putative [Ricinus communis] |
| 20 |
Hb_000364_030 |
0.0746771348 |
- |
- |
RNA binding protein, putative [Ricinus communis] |