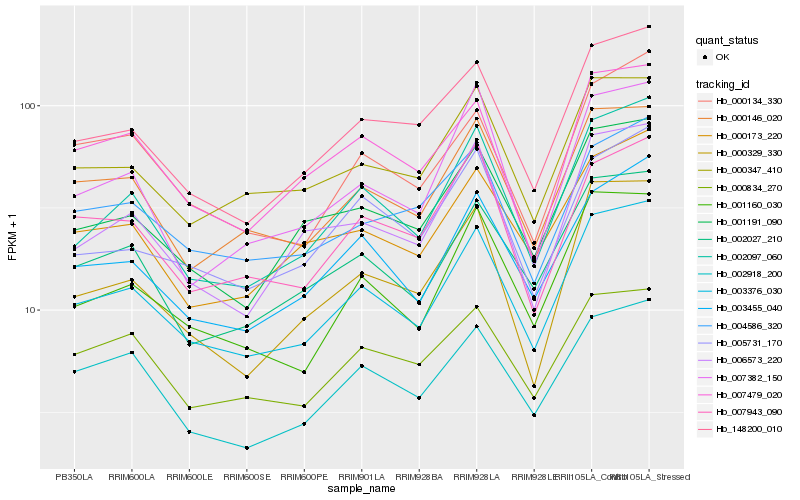
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003376_030 |
0.0 |
- |
- |
PREDICTED: ribonuclease P protein subunit p29 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003455_040 |
0.0525790818 |
- |
- |
hypothetical protein JCGZ_17270 [Jatropha curcas] |
| 3 |
Hb_002097_060 |
0.0538528878 |
- |
- |
PREDICTED: 50S ribosomal protein L9, chloroplastic-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_148200_010 |
0.0572426372 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 5 |
Hb_000146_020 |
0.0575814087 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001160_030 |
0.0580352501 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein POPTR_0018s05470g [Populus trichocarpa] |
| 7 |
Hb_002027_210 |
0.0598662216 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 8 |
Hb_001191_090 |
0.064529682 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 9 |
Hb_000347_410 |
0.0660043549 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 10 |
Hb_002918_200 |
0.0676192878 |
- |
- |
- |
| 11 |
Hb_000173_220 |
0.0692828699 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 12 |
Hb_006573_220 |
0.0715850667 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 13 |
Hb_007479_020 |
0.072840845 |
- |
- |
PREDICTED: CMP-sialic acid transporter 5 [Jatropha curcas] |
| 14 |
Hb_005731_170 |
0.0732237875 |
- |
- |
hypothetical protein PRUPE_ppa011750mg [Prunus persica] |
| 15 |
Hb_007943_090 |
0.0740733112 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 16 |
Hb_004586_320 |
0.074308459 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 17 |
Hb_000329_330 |
0.0743379929 |
- |
- |
PREDICTED: vesicle-associated protein 1-2-like [Jatropha curcas] |
| 18 |
Hb_000134_330 |
0.0743571898 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm7 [Elaeis guineensis] |
| 19 |
Hb_000834_270 |
0.0780428015 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007382_150 |
0.0788636709 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |