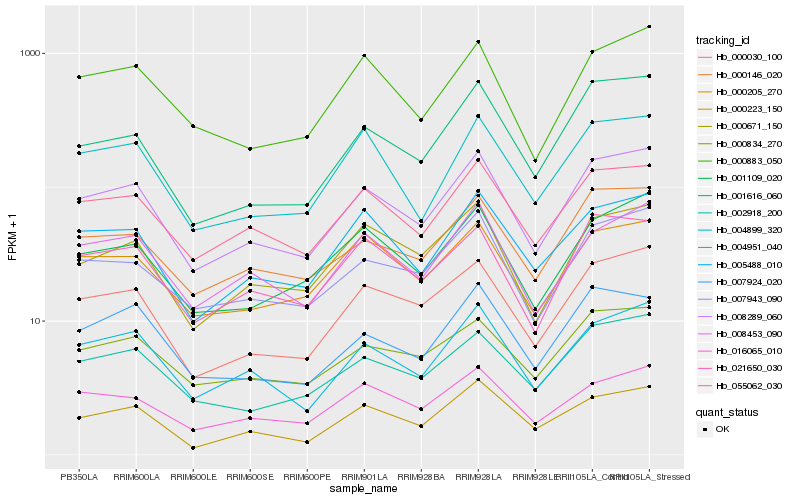
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008289_060 |
0.0 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000030_100 |
0.056397388 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |
| 3 |
Hb_004951_040 |
0.0565097096 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g31790-like [Jatropha curcas] |
| 4 |
Hb_055062_030 |
0.0627894122 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 10b-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_007943_090 |
0.0640376679 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 6 |
Hb_000146_020 |
0.0649740661 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002918_200 |
0.0687804676 |
- |
- |
- |
| 8 |
Hb_005488_010 |
0.0706517949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001109_020 |
0.0709267998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_008453_090 |
0.071338217 |
- |
- |
hypothetical protein JCGZ_08177 [Jatropha curcas] |
| 11 |
Hb_000223_150 |
0.0731549577 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Populus euphratica] |
| 12 |
Hb_021650_030 |
0.0739163958 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 13 |
Hb_000834_270 |
0.0742670032 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 14 |
Hb_016065_010 |
0.0748805746 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08305 [Jatropha curcas] |
| 15 |
Hb_000205_270 |
0.0775016746 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000671_150 |
0.0779941423 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 17 |
Hb_000883_050 |
0.0789400311 |
- |
- |
PREDICTED: 60S ribosomal protein L37a-like, partial [Solanum tuberosum] |
| 18 |
Hb_007924_020 |
0.0790417696 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 19 |
Hb_001616_060 |
0.0791191356 |
- |
- |
PREDICTED: DNA-directed RNA polymerase subunit 10-like protein [Jatropha curcas] |
| 20 |
Hb_004899_320 |
0.0803077175 |
- |
- |
PREDICTED: oligouridylate-binding protein 1-like [Jatropha curcas] |