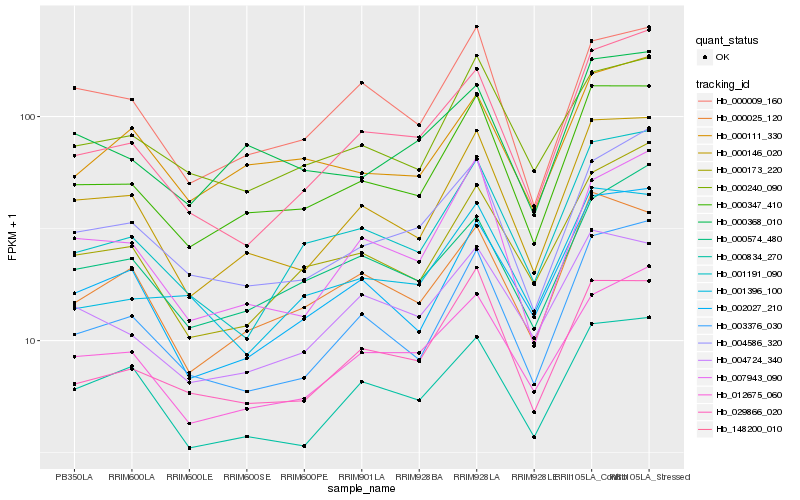
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_410 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 2 |
Hb_000146_020 |
0.050105788 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 3 |
Hb_029866_020 |
0.0616886842 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 [Jatropha curcas] |
| 4 |
Hb_000025_120 |
0.0655737443 |
- |
- |
PREDICTED: uncharacterized protein LOC105628946 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003376_030 |
0.0660043549 |
- |
- |
PREDICTED: ribonuclease P protein subunit p29 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004586_320 |
0.0710695617 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 7 |
Hb_000240_090 |
0.071116801 |
- |
- |
Rab5 [Hevea brasiliensis] |
| 8 |
Hb_012675_060 |
0.0723726146 |
- |
- |
hypothetical protein PRUPE_ppa007715mg [Prunus persica] |
| 9 |
Hb_001191_090 |
0.0747469873 |
- |
- |
PREDICTED: uncharacterized protein LOC105643880 [Jatropha curcas] |
| 10 |
Hb_004724_340 |
0.0754927861 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 11 |
Hb_000368_010 |
0.0758128935 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 18 [Jatropha curcas] |
| 12 |
Hb_007943_090 |
0.0762189174 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 13 |
Hb_000173_220 |
0.0767544864 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein At5g19025 [Jatropha curcas] |
| 14 |
Hb_002027_210 |
0.0768401895 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 15 |
Hb_000009_160 |
0.0772216486 |
- |
- |
RUB1 conjugating enzyme 1 isoform 2 [Theobroma cacao] |
| 16 |
Hb_000574_480 |
0.078306527 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 17 |
Hb_000834_270 |
0.0803590526 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000111_330 |
0.0804514947 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 [Jatropha curcas] |
| 19 |
Hb_148200_010 |
0.0809375867 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 20 |
Hb_001396_100 |
0.081037679 |
- |
- |
DNA binding protein, putative [Ricinus communis] |