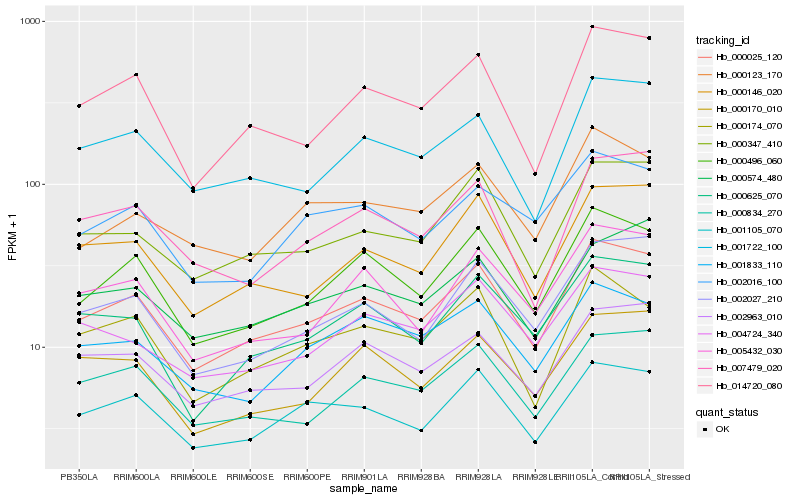
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628946 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000496_060 |
0.0542443776 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH3 [Jatropha curcas] |
| 3 |
Hb_014720_080 |
0.062676287 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 4 |
Hb_000347_410 |
0.0655737443 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 5 |
Hb_001105_070 |
0.0676099083 |
- |
- |
PREDICTED: uncharacterized protein LOC105632829 [Jatropha curcas] |
| 6 |
Hb_000146_020 |
0.0697999292 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000834_270 |
0.0701618087 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002963_010 |
0.0706016572 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002027_210 |
0.0706152578 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 10 |
Hb_001722_100 |
0.0709197075 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000625_070 |
0.07370686 |
transcription factor |
TF Family: CSD |
PREDICTED: cold shock domain-containing protein 3 [Jatropha curcas] |
| 12 |
Hb_005432_030 |
0.0756399478 |
- |
- |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 [Gossypium raimondii] |
| 13 |
Hb_000174_070 |
0.0788671764 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_004724_340 |
0.0797565553 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 15 |
Hb_001833_110 |
0.0819427792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002016_100 |
0.0823890306 |
- |
- |
Small nuclear ribonucleoprotein SM D3, putative [Ricinus communis] |
| 17 |
Hb_007479_020 |
0.0826406617 |
- |
- |
PREDICTED: CMP-sialic acid transporter 5 [Jatropha curcas] |
| 18 |
Hb_000170_010 |
0.0829113547 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000574_480 |
0.0837913052 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 20 |
Hb_000123_170 |
0.0838492554 |
- |
- |
Rab3 [Hevea brasiliensis] |