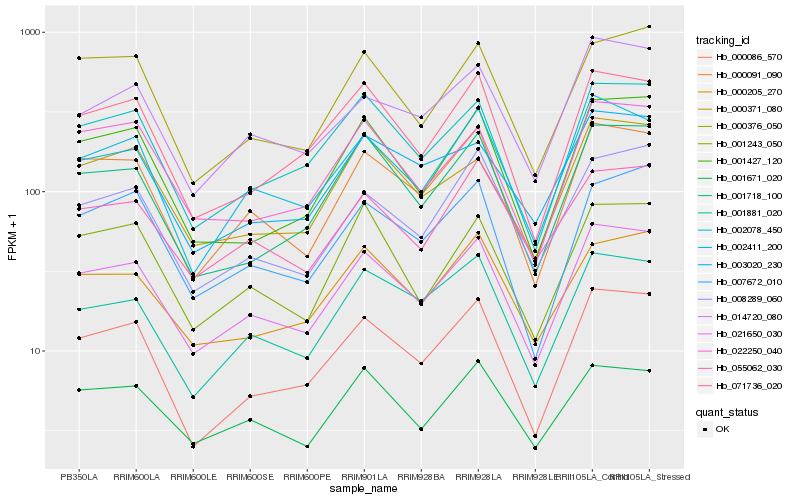
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021650_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 2 |
Hb_000091_090 |
0.050869995 |
- |
- |
hypothetical protein JCGZ_09031 [Jatropha curcas] |
| 3 |
Hb_002411_200 |
0.055591233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000086_570 |
0.0591683073 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 5 |
Hb_002078_450 |
0.0621804264 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 6 |
Hb_014720_080 |
0.0671514756 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 7 |
Hb_001881_020 |
0.0675860271 |
- |
- |
clathrin binding protein, putative [Ricinus communis] |
| 8 |
Hb_001671_020 |
0.0697908738 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08510 [Jatropha curcas] |
| 9 |
Hb_000205_270 |
0.0703280509 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001718_100 |
0.0727254689 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 11 |
Hb_003020_230 |
0.0728226486 |
- |
- |
DNA-directed RNA polymerase II subunit RPB7 [Glycine max] |
| 12 |
Hb_071736_020 |
0.0732684438 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 13 |
Hb_007672_010 |
0.0734664523 |
- |
- |
PREDICTED: uncharacterized protein LOC105639669 isoform X1 [Jatropha curcas] |
| 14 |
Hb_055062_030 |
0.0735362384 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 10b-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000371_080 |
0.0738077978 |
- |
- |
ubiquitin-conjugating enzyme m, putative [Ricinus communis] |
| 16 |
Hb_008289_060 |
0.0739163958 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_022250_040 |
0.0740655782 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001427_120 |
0.0744090263 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 19 |
Hb_000376_050 |
0.0745541464 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 20 |
Hb_001243_050 |
0.0754838439 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |