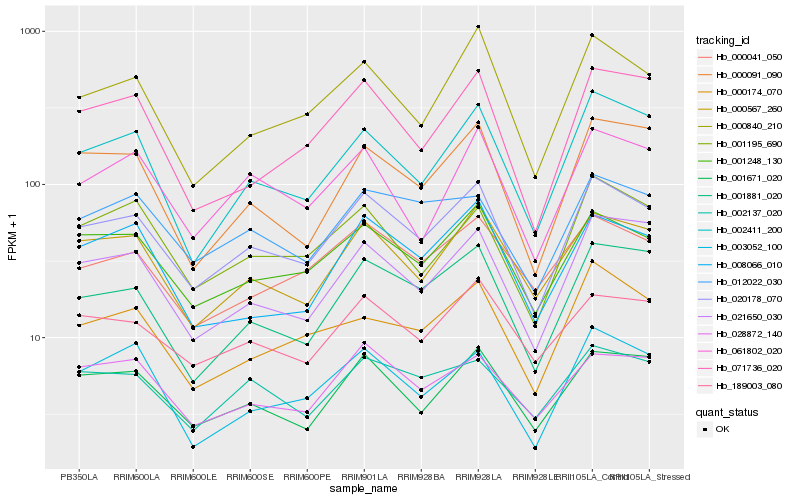
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020178_070 |
0.0 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 2 |
Hb_001248_130 |
0.0645967868 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000567_260 |
0.0745449349 |
- |
- |
BnaA09g41440D [Brassica napus] |
| 4 |
Hb_001671_020 |
0.0762112693 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08510 [Jatropha curcas] |
| 5 |
Hb_001881_020 |
0.0781994048 |
- |
- |
clathrin binding protein, putative [Ricinus communis] |
| 6 |
Hb_021650_030 |
0.0800381022 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 7 |
Hb_002411_200 |
0.0815491199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000840_210 |
0.0833722153 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 9 |
Hb_189003_080 |
0.0849962624 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 10 |
Hb_001195_690 |
0.0879645502 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185 [Jatropha curcas] |
| 11 |
Hb_012022_030 |
0.0907206689 |
transcription factor |
TF Family: Alfin-like |
phd/F-box containing protein, putative [Ricinus communis] |
| 12 |
Hb_000174_070 |
0.0910703559 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000041_050 |
0.0912638225 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 14 |
Hb_061802_020 |
0.0926112473 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 15 |
Hb_028872_140 |
0.0928360984 |
- |
- |
PREDICTED: uncharacterized protein LOC105644767 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000091_090 |
0.0929770179 |
- |
- |
hypothetical protein JCGZ_09031 [Jatropha curcas] |
| 17 |
Hb_008066_010 |
0.0932929733 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit alpha-like [Jatropha curcas] |
| 18 |
Hb_002137_020 |
0.0933464966 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_003052_100 |
0.0956147209 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_071736_020 |
0.0959646583 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |