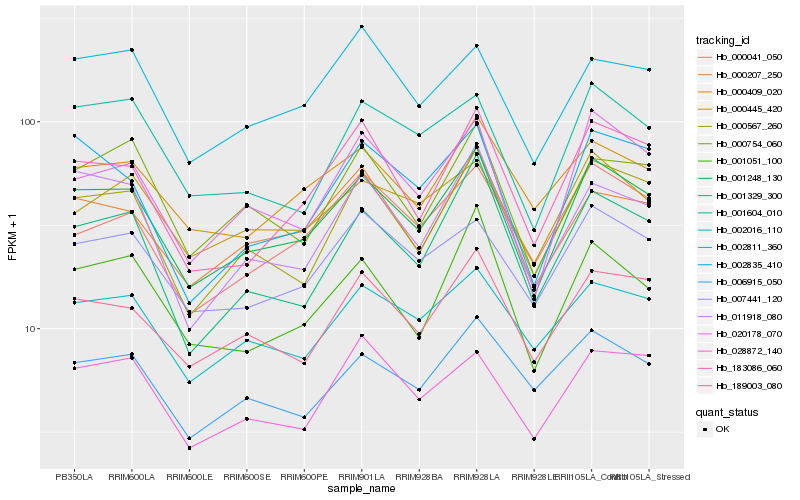
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001248_130 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 2 |
Hb_020178_070 |
0.0645967868 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 3 |
Hb_000567_260 |
0.0662203998 |
- |
- |
BnaA09g41440D [Brassica napus] |
| 4 |
Hb_189003_080 |
0.0679571368 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 5 |
Hb_007441_120 |
0.0703610264 |
- |
- |
PREDICTED: dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase isoform X2 [Jatropha curcas] |
| 6 |
Hb_000207_250 |
0.0713339523 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 7 |
Hb_002016_110 |
0.0726315947 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 8 |
Hb_001051_100 |
0.0752220981 |
transcription factor |
TF Family: ARF |
Auxin response factor, putative [Ricinus communis] |
| 9 |
Hb_183086_060 |
0.0752733009 |
desease resistance |
Gene Name: AAA |
PREDICTED: ATPase family AAA domain-containing protein 1-B [Jatropha curcas] |
| 10 |
Hb_011918_080 |
0.0758688737 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000409_020 |
0.0781595594 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 12 |
Hb_001329_300 |
0.0783359908 |
- |
- |
PREDICTED: L-cysteine desulfhydrase [Jatropha curcas] |
| 13 |
Hb_000754_060 |
0.0792981438 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001604_010 |
0.0796671399 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 15 |
Hb_000445_420 |
0.0798846435 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 16 |
Hb_028872_140 |
0.0805538632 |
- |
- |
PREDICTED: uncharacterized protein LOC105644767 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000041_050 |
0.0806199288 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 18 |
Hb_002835_410 |
0.0806934996 |
- |
- |
clathrin coat assembly protein ap-1, putative [Ricinus communis] |
| 19 |
Hb_006915_050 |
0.0809571617 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002811_360 |
0.0814098291 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |