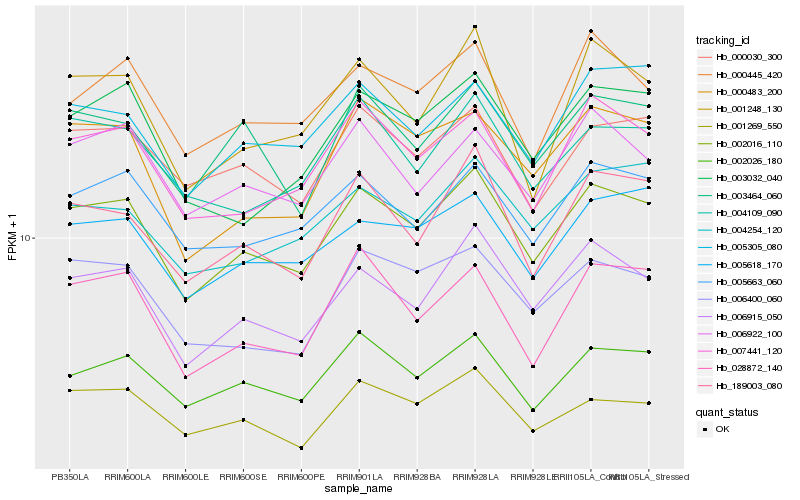
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002016_110 |
0.0 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 2 |
Hb_006915_050 |
0.0494848228 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002026_180 |
0.0561160848 |
- |
- |
PREDICTED: uncharacterized protein LOC105645805 [Jatropha curcas] |
| 4 |
Hb_007441_120 |
0.0603427098 |
- |
- |
PREDICTED: dol-P-Man:Man(7)GlcNAc(2)-PP-Dol alpha-1,6-mannosyltransferase isoform X2 [Jatropha curcas] |
| 5 |
Hb_005663_060 |
0.0618487932 |
- |
- |
PREDICTED: uncharacterized protein LOC105770600 [Gossypium raimondii] |
| 6 |
Hb_189003_080 |
0.0620872421 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 7 |
Hb_000030_300 |
0.0645860853 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 8 |
Hb_005618_170 |
0.0664013163 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 9 |
Hb_003464_060 |
0.0673672423 |
- |
- |
PREDICTED: F-box protein PP2-A15 [Jatropha curcas] |
| 10 |
Hb_000483_200 |
0.0675388732 |
- |
- |
Phospho-2-dehydro-3-deoxyheptonate aldolase 1, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_006400_060 |
0.0687200744 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 31 isoform X1 [Jatropha curcas] |
| 12 |
Hb_006922_100 |
0.068909683 |
- |
- |
PREDICTED: uncharacterized protein LOC105645470 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000445_420 |
0.06905119 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 14 |
Hb_004109_090 |
0.0692259579 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_001269_550 |
0.0706914493 |
- |
- |
hypothetical protein POPTR_0001s03510g [Populus trichocarpa] |
| 16 |
Hb_005305_080 |
0.0711435387 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003032_040 |
0.072320707 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: zinc finger CCCH domain-containing protein 25 [Jatropha curcas] |
| 18 |
Hb_001248_130 |
0.0726315947 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004254_120 |
0.0730702762 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_028872_140 |
0.0740132422 |
- |
- |
PREDICTED: uncharacterized protein LOC105644767 isoform X1 [Jatropha curcas] |