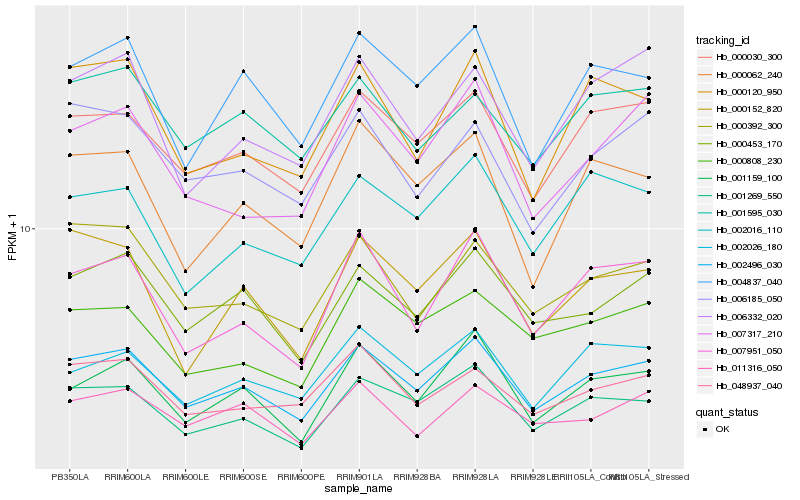
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002496_030 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g69350, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000030_300 |
0.0518257976 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 3 |
Hb_000453_170 |
0.0528557498 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_001269_550 |
0.0543193295 |
- |
- |
hypothetical protein POPTR_0001s03510g [Populus trichocarpa] |
| 5 |
Hb_004837_040 |
0.06326827 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1-like [Prunus mume] |
| 6 |
Hb_007951_050 |
0.065536018 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000392_300 |
0.0697704456 |
- |
- |
PREDICTED: RINT1-like protein MAG2 [Jatropha curcas] |
| 8 |
Hb_000120_950 |
0.0705131689 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 9 |
Hb_002026_180 |
0.0714327519 |
- |
- |
PREDICTED: uncharacterized protein LOC105645805 [Jatropha curcas] |
| 10 |
Hb_011316_050 |
0.0725036466 |
- |
- |
hypothetical protein CICLE_v10004643mg [Citrus clementina] |
| 11 |
Hb_006185_050 |
0.0725501297 |
- |
- |
hypothetical protein POPTR_0001s20590g [Populus trichocarpa] |
| 12 |
Hb_001159_100 |
0.0727617019 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g22760 [Jatropha curcas] |
| 13 |
Hb_048937_040 |
0.0729347791 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 14 |
Hb_001595_030 |
0.0733970091 |
- |
- |
PREDICTED: nuclear inhibitor of protein phosphatase 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_006332_020 |
0.074068336 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 16 |
Hb_000152_820 |
0.0744001211 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g64320, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000808_230 |
0.0746643973 |
- |
- |
PREDICTED: peptide chain release factor PrfB2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002016_110 |
0.0748200202 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 19 |
Hb_000062_240 |
0.0748867281 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 20 |
Hb_007317_210 |
0.0754970603 |
- |
- |
PREDICTED: probable calcium-binding protein CML48 [Jatropha curcas] |