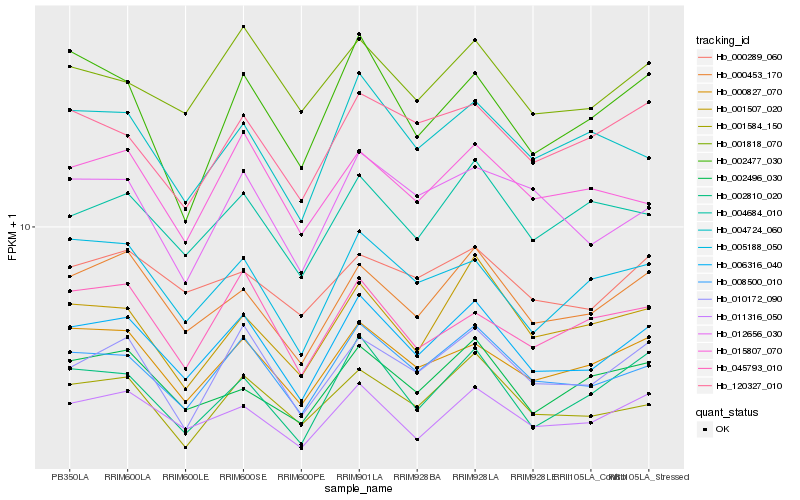
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006316_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g16860 [Jatropha curcas] |
| 2 |
Hb_008500_010 |
0.0439028131 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17210 [Jatropha curcas] |
| 3 |
Hb_000453_170 |
0.0603470728 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_011316_050 |
0.0632448451 |
- |
- |
hypothetical protein CICLE_v10004643mg [Citrus clementina] |
| 5 |
Hb_000827_070 |
0.0675707004 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 6 |
Hb_010172_090 |
0.0743424186 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_005188_050 |
0.0754719398 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 2B, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_001584_150 |
0.0771245827 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 9 |
Hb_004684_010 |
0.0798954253 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 10 |
Hb_004724_060 |
0.0810574361 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001818_070 |
0.0814914086 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 12 |
Hb_002496_030 |
0.0837430994 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g69350, mitochondrial [Jatropha curcas] |
| 13 |
Hb_012656_030 |
0.0840069243 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001507_020 |
0.0848760738 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14170 [Jatropha curcas] |
| 15 |
Hb_015807_070 |
0.0857369819 |
- |
- |
PREDICTED: autophagy-related protein 18f [Jatropha curcas] |
| 16 |
Hb_120327_010 |
0.0859499603 |
- |
- |
PREDICTED: uncharacterized protein LOC105650959 [Jatropha curcas] |
| 17 |
Hb_002477_030 |
0.0859815249 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105631404 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000289_060 |
0.0867619026 |
- |
- |
PREDICTED: uncharacterized protein At1g18480 [Jatropha curcas] |
| 19 |
Hb_002810_020 |
0.0873708895 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g25360 [Jatropha curcas] |
| 20 |
Hb_045793_010 |
0.087496554 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Pyrus x bretschneideri] |