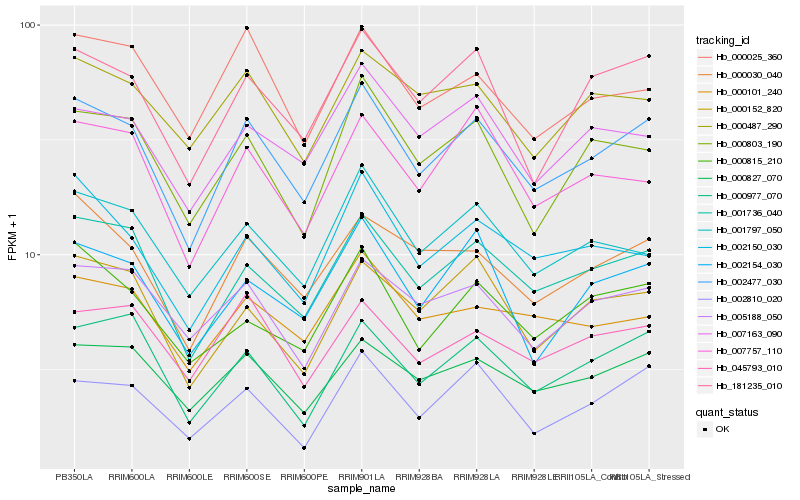
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002477_030 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105631404 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001736_040 |
0.0553184657 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Jatropha curcas] |
| 3 |
Hb_000827_070 |
0.063159166 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 4 |
Hb_181235_010 |
0.0673392857 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105645554 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000030_040 |
0.0734341418 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001797_050 |
0.0772801344 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_005188_050 |
0.0800363624 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 2B, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 8 |
Hb_002154_030 |
0.0802911803 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_000803_190 |
0.0807581849 |
- |
- |
PREDICTED: THUMP domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007163_090 |
0.0819669571 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 11 |
Hb_000977_070 |
0.0823540344 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_007757_110 |
0.0824329784 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 13 |
Hb_000152_820 |
0.0829376241 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g64320, mitochondrial [Jatropha curcas] |
| 14 |
Hb_002810_020 |
0.0833278302 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g25360 [Jatropha curcas] |
| 15 |
Hb_000101_240 |
0.0836691388 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 16 |
Hb_045793_010 |
0.0848658676 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Pyrus x bretschneideri] |
| 17 |
Hb_000487_290 |
0.0851033015 |
- |
- |
ribonucleic acid binding protein S1, putative [Ricinus communis] |
| 18 |
Hb_002150_030 |
0.0853118409 |
- |
- |
PREDICTED: protein SPA1-RELATED 2 [Jatropha curcas] |
| 19 |
Hb_000025_360 |
0.0853186308 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000815_210 |
0.0856085319 |
- |
- |
uncoupling protein 3, partial [Manihot esculenta] |