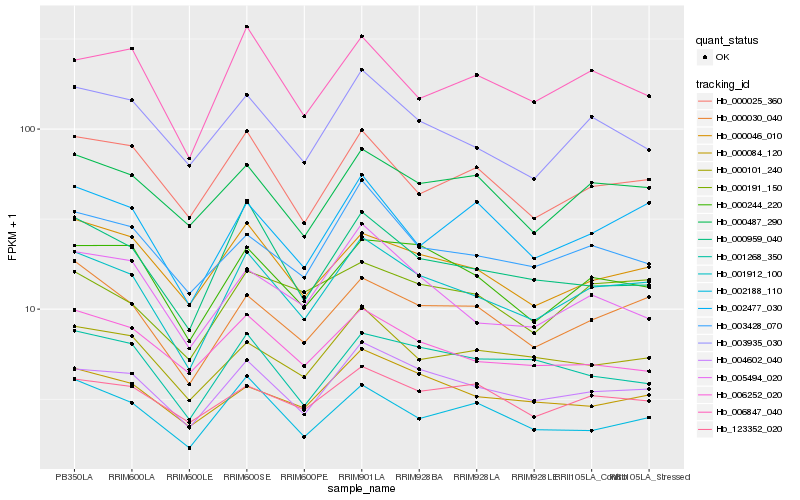
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001912_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636609 [Jatropha curcas] |
| 2 |
Hb_004602_040 |
0.0555110481 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 3 |
Hb_000030_040 |
0.0714996304 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000244_220 |
0.074047916 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000046_010 |
0.0818534933 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 6 |
Hb_003935_030 |
0.0827077506 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 7 |
Hb_000487_290 |
0.0829449221 |
- |
- |
ribonucleic acid binding protein S1, putative [Ricinus communis] |
| 8 |
Hb_000084_120 |
0.0857294899 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g36240 [Jatropha curcas] |
| 9 |
Hb_000101_240 |
0.0866985068 |
desease resistance |
Gene Name: NB-ARC |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 10 |
Hb_003428_070 |
0.0875861326 |
- |
- |
DEAD-box ATP-dependent RNA helicase 46 -like protein [Gossypium arboreum] |
| 11 |
Hb_000959_040 |
0.088896665 |
- |
- |
PREDICTED: uncharacterized protein LOC105636570 [Jatropha curcas] |
| 12 |
Hb_002477_030 |
0.089304603 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105631404 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000025_360 |
0.0900983644 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000191_150 |
0.0901404546 |
- |
- |
PREDICTED: uncharacterized protein LOC105637951 [Jatropha curcas] |
| 15 |
Hb_002188_110 |
0.0906225391 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 16 |
Hb_006847_040 |
0.0920739015 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 1 [Jatropha curcas] |
| 17 |
Hb_005494_020 |
0.0944105269 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 18 |
Hb_123352_020 |
0.0948457677 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560 [Jatropha curcas] |
| 19 |
Hb_001268_350 |
0.0967477967 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105115846 [Populus euphratica] |
| 20 |
Hb_006252_020 |
0.0969614687 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |