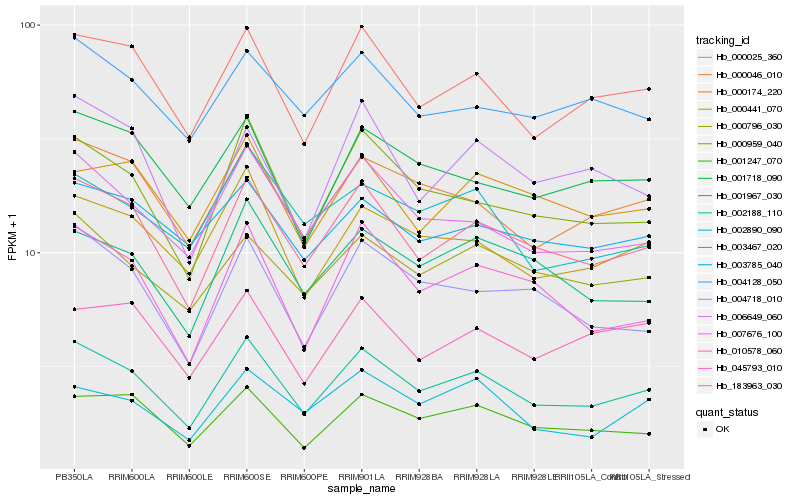
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002188_110 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 2 |
Hb_183963_030 |
0.042142318 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000025_360 |
0.0597012123 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000959_040 |
0.0607405628 |
- |
- |
PREDICTED: uncharacterized protein LOC105636570 [Jatropha curcas] |
| 5 |
Hb_007676_100 |
0.06610184 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 6 |
Hb_000441_070 |
0.0694002408 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001247_070 |
0.0697151922 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g56690, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000046_010 |
0.0699566492 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |
| 9 |
Hb_010578_060 |
0.0718441683 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 14 [Jatropha curcas] |
| 10 |
Hb_004718_010 |
0.0784008794 |
- |
- |
PREDICTED: uncharacterized protein LOC105638290 [Jatropha curcas] |
| 11 |
Hb_001967_030 |
0.0798134191 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |
| 12 |
Hb_000796_030 |
0.0817146331 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32450, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001718_090 |
0.0817235887 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 97 [Jatropha curcas] |
| 14 |
Hb_004128_050 |
0.0870868245 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_003467_020 |
0.0873251242 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 13-like [Jatropha curcas] |
| 16 |
Hb_006649_060 |
0.0875798921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000174_220 |
0.0892284653 |
- |
- |
cohesin subunit rad21, putative [Ricinus communis] |
| 18 |
Hb_002890_090 |
0.0895781273 |
- |
- |
Auxin response factor 1 [Glycine soja] |
| 19 |
Hb_045793_010 |
0.0895782902 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Pyrus x bretschneideri] |
| 20 |
Hb_003785_040 |
0.0895909518 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |