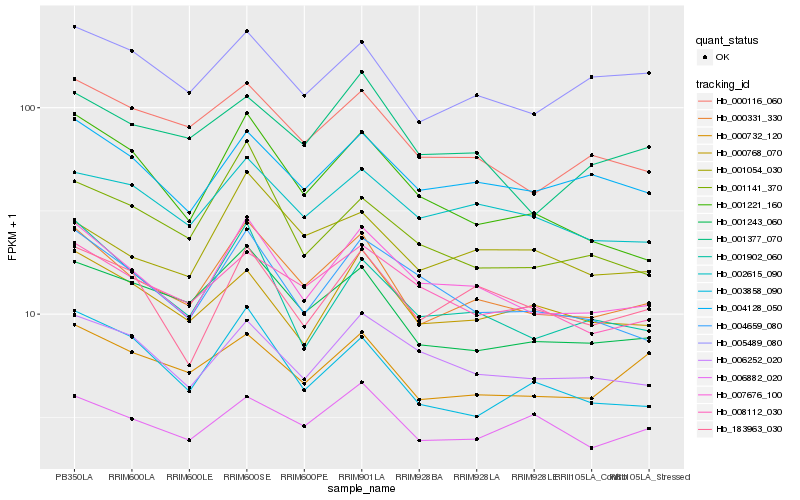
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_330 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 2 |
Hb_007676_100 |
0.0609547277 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 3 |
Hb_001243_060 |
0.0671802676 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 4 |
Hb_000116_060 |
0.076540019 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 5 |
Hb_004128_050 |
0.0812195749 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_005489_080 |
0.0850083875 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002615_090 |
0.0851168039 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_008112_030 |
0.0853528015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000732_120 |
0.0854393648 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001377_070 |
0.087924018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001054_030 |
0.0886786083 |
transcription factor |
TF Family: NF-X1 |
PREDICTED: NF-X1-type zinc finger protein NFXL1 [Jatropha curcas] |
| 12 |
Hb_003858_090 |
0.0908015543 |
- |
- |
Filamin-A-interacting protein, putative [Ricinus communis] |
| 13 |
Hb_006882_020 |
0.0916300131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004659_080 |
0.093016732 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 15 |
Hb_183963_030 |
0.0934429856 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_006252_020 |
0.0939646237 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000768_070 |
0.0943522737 |
transcription factor |
TF Family: SNF2 |
PREDICTED: TATA-binding protein-associated factor BTAF1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001221_160 |
0.0949007668 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 19 |
Hb_001141_370 |
0.0949387482 |
- |
- |
PREDICTED: uncharacterized protein LOC105631986 [Jatropha curcas] |
| 20 |
Hb_001902_060 |
0.0952877868 |
- |
- |
PREDICTED: U-box domain-containing protein 32 isoform X3 [Jatropha curcas] |