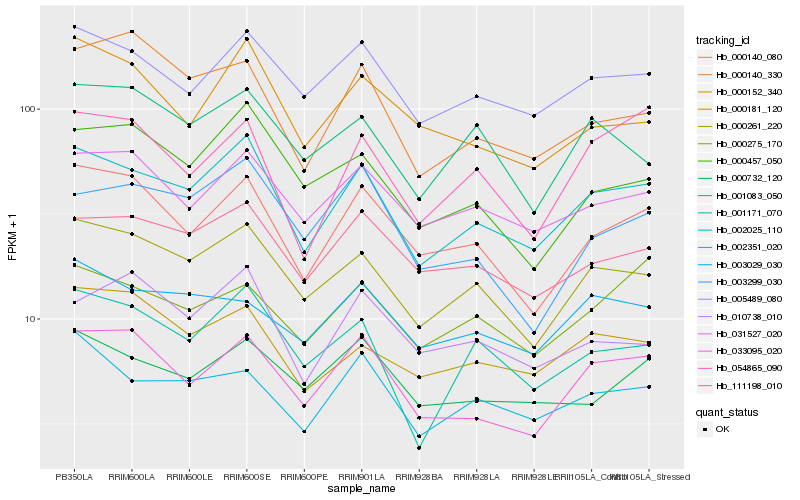
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002025_110 |
0.0 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_000261_220 |
0.062914894 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 3 |
Hb_005489_080 |
0.0694333188 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_111198_010 |
0.078185759 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_033095_020 |
0.0802575851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000140_080 |
0.0803917196 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_031527_020 |
0.0807325843 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 8 |
Hb_001171_070 |
0.0821833214 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_002351_020 |
0.0871664683 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000457_050 |
0.0891051261 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 11 |
Hb_000732_120 |
0.0901566065 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000181_120 |
0.09295583 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 13 |
Hb_000152_340 |
0.0958473128 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 14 |
Hb_010738_010 |
0.0972306247 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 15 |
Hb_003029_030 |
0.0988707819 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001083_050 |
0.0993988261 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 17 |
Hb_003299_030 |
0.1005728502 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000275_170 |
0.100725764 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 19 |
Hb_054865_090 |
0.1011328813 |
- |
- |
GRF1-INTERACTING FACTOR 2 family protein [Populus trichocarpa] |
| 20 |
Hb_000140_330 |
0.1017339862 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |