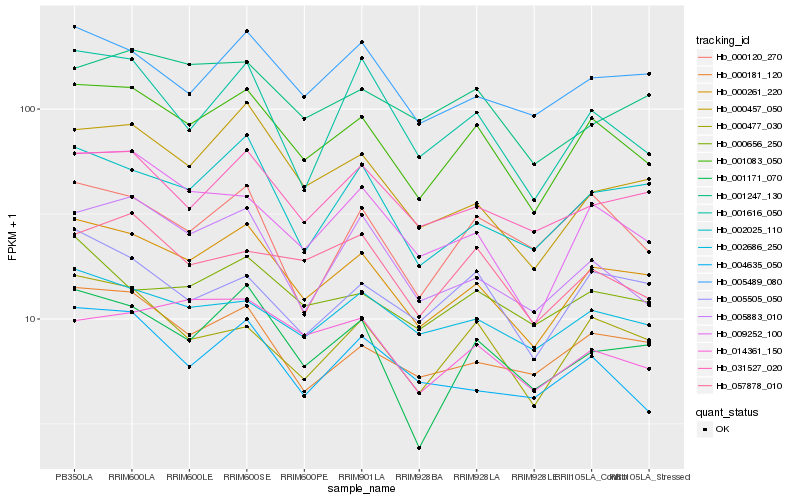
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001083_050 |
0.0 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 2 |
Hb_000261_220 |
0.0506460438 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 3 |
Hb_000477_030 |
0.0790684163 |
- |
- |
PREDICTED: boron transporter 4-like [Jatropha curcas] |
| 4 |
Hb_009252_100 |
0.0831051242 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 5 |
Hb_057878_010 |
0.0836239574 |
- |
- |
hypothetical protein VITISV_036304 [Vitis vinifera] |
| 6 |
Hb_005883_010 |
0.0888448726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001171_070 |
0.0891880119 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002686_250 |
0.0903183845 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 9 |
Hb_000181_120 |
0.0906266963 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 10 |
Hb_031527_020 |
0.0912730366 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 11 |
Hb_005505_050 |
0.0930797365 |
- |
- |
PREDICTED: uncharacterized protein LOC105639309 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000457_050 |
0.0936009461 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 13 |
Hb_005489_080 |
0.0944236387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004635_050 |
0.0967162866 |
- |
- |
nucleolar phosphoprotein, putative [Ricinus communis] |
| 15 |
Hb_000656_250 |
0.0975825985 |
- |
- |
hypothetical protein JCGZ_24680 [Jatropha curcas] |
| 16 |
Hb_000120_270 |
0.0977681229 |
- |
- |
PREDICTED: histone deacetylase 8 [Jatropha curcas] |
| 17 |
Hb_001616_050 |
0.0979854619 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 18 |
Hb_014361_150 |
0.099145254 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 19 |
Hb_002025_110 |
0.0993988261 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_001247_130 |
0.1000141902 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |