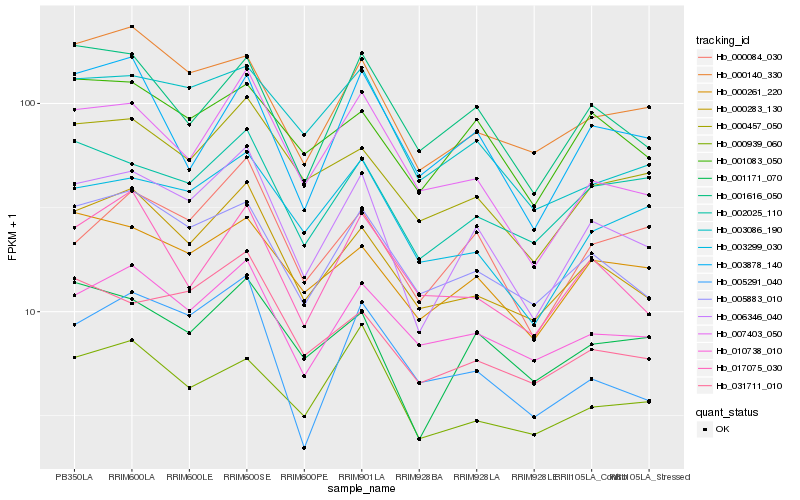
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006346_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_20335 [Jatropha curcas] |
| 2 |
Hb_000283_130 |
0.0923715441 |
- |
- |
PREDICTED: PRA1 family protein E-like [Populus euphratica] |
| 3 |
Hb_001171_070 |
0.0940073847 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit alpha 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_005883_010 |
0.0980025067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007403_050 |
0.1008978754 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 6 |
Hb_000084_030 |
0.1010525721 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000457_050 |
0.1024275042 |
- |
- |
PREDICTED: tether containing UBX domain for GLUT4 [Jatropha curcas] |
| 8 |
Hb_002025_110 |
0.1041022187 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: SWI/SNF complex component SNF12 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_000261_220 |
0.1048452051 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 10 |
Hb_003299_030 |
0.106278387 |
- |
- |
PREDICTED: uncharacterized protein At1g27050 isoform X1 [Jatropha curcas] |
| 11 |
Hb_010738_010 |
0.1064953901 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 12 |
Hb_001083_050 |
0.1065230532 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 13 |
Hb_001616_050 |
0.1093159775 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 14 |
Hb_000140_330 |
0.1103877754 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 15 |
Hb_031711_010 |
0.1104211688 |
- |
- |
PREDICTED: CDPK-related kinase 4-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003086_190 |
0.1143402813 |
- |
- |
PREDICTED: uncharacterized protein LOC105635794 [Jatropha curcas] |
| 17 |
Hb_005291_040 |
0.1196826271 |
- |
- |
PREDICTED: gamma-gliadin [Jatropha curcas] |
| 18 |
Hb_003878_140 |
0.1227856328 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 19 |
Hb_000939_060 |
0.1230050481 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 20 |
Hb_017075_030 |
0.1243374799 |
- |
- |
helicase, putative [Ricinus communis] |