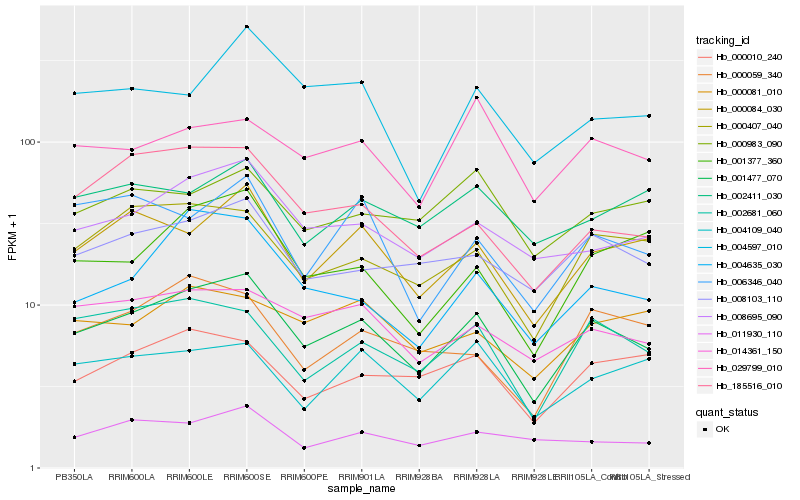
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001477_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000084_030 |
0.1059245482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008695_090 |
0.1064650772 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 4 |
Hb_000407_040 |
0.1093396716 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_008103_110 |
0.11203231 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_002681_060 |
0.1139026783 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 7 |
Hb_014361_150 |
0.1156626743 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 8 |
Hb_004635_030 |
0.1163713026 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 9 |
Hb_185516_010 |
0.1217622489 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 10 |
Hb_001377_360 |
0.123858925 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 11 |
Hb_000010_240 |
0.1260028969 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 12 |
Hb_011930_110 |
0.1292796647 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g59720, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000059_340 |
0.1308346316 |
transcription factor |
TF Family: SOH1 |
uncharacterized protein LOC100527879 [Glycine max] |
| 14 |
Hb_004597_010 |
0.1329987165 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor MYC2 [Jatropha curcas] |
| 15 |
Hb_004109_040 |
0.1332095608 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g53600, mitochondrial [Jatropha curcas] |
| 16 |
Hb_006346_040 |
0.1336839883 |
- |
- |
hypothetical protein JCGZ_20335 [Jatropha curcas] |
| 17 |
Hb_000081_010 |
0.1342009617 |
- |
- |
hypothetical protein B456_012G056000 [Gossypium raimondii] |
| 18 |
Hb_029799_010 |
0.1343461988 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_000983_090 |
0.1370375813 |
- |
- |
PREDICTED: mitogen-activated protein kinase homolog NTF3 [Jatropha curcas] |
| 20 |
Hb_002411_030 |
0.1378721933 |
transcription factor |
TF Family: NF-YC |
PREDICTED: dr1-associated corepressor homolog [Jatropha curcas] |