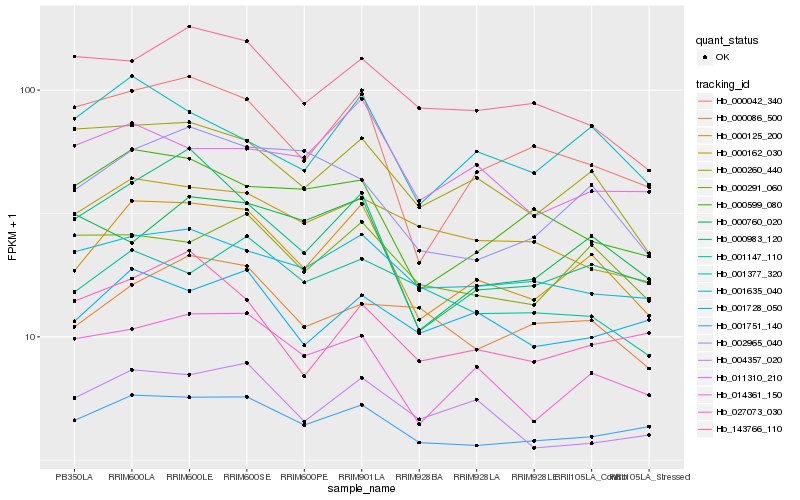
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000125_200 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: probable transcription factor PosF21 [Jatropha curcas] |
| 2 |
Hb_014361_150 |
0.0820170154 |
- |
- |
PREDICTED: uncharacterized protein LOC105639827 [Jatropha curcas] |
| 3 |
Hb_000983_120 |
0.090116308 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000260_440 |
0.0910004176 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 5 |
Hb_000042_340 |
0.0918714229 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 6 |
Hb_001377_320 |
0.0930053301 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 7 |
Hb_000291_060 |
0.0972495315 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_004357_020 |
0.0986168895 |
- |
- |
PREDICTED: uncharacterized protein LOC105637355 [Jatropha curcas] |
| 9 |
Hb_001635_040 |
0.1018422032 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B beta isoform-like isoform X2 [Gossypium raimondii] |
| 10 |
Hb_000599_080 |
0.1020985105 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 11 |
Hb_002965_040 |
0.103361156 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 12 |
Hb_001751_140 |
0.103933825 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |
| 13 |
Hb_143766_110 |
0.1061082494 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 14 |
Hb_011310_210 |
0.1062018655 |
- |
- |
PREDICTED: serine/threonine-protein kinase PRP4 homolog [Jatropha curcas] |
| 15 |
Hb_000162_030 |
0.106431689 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_001728_050 |
0.1074149076 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 17 |
Hb_001147_110 |
0.1076728187 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 18 |
Hb_000086_500 |
0.1098764646 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_000760_020 |
0.1103377122 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_027073_030 |
0.1122575972 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |