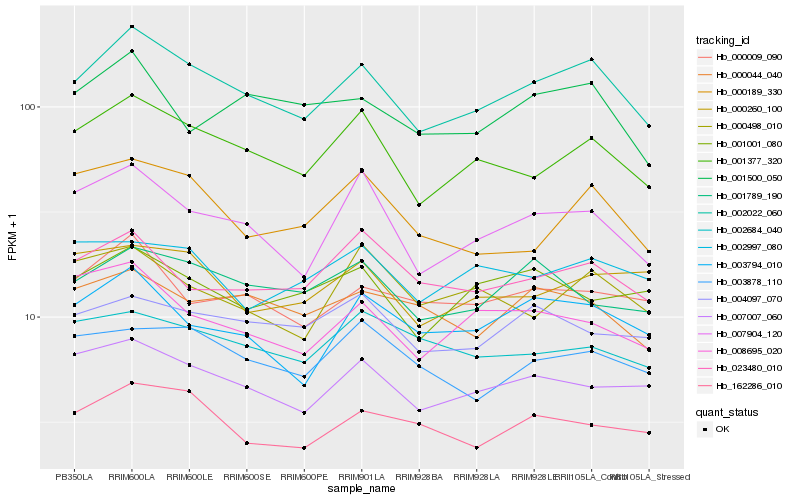
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002022_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637912 [Jatropha curcas] |
| 2 |
Hb_001377_320 |
0.0651794873 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |
| 3 |
Hb_007904_120 |
0.0767787346 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007007_060 |
0.0814296213 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_003794_010 |
0.0871798762 |
- |
- |
PREDICTED: uncharacterized protein LOC105641013 [Jatropha curcas] |
| 6 |
Hb_000009_090 |
0.089436875 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 7 |
Hb_162286_010 |
0.0917484137 |
- |
- |
PREDICTED: uncharacterized protein LOC105642699 [Jatropha curcas] |
| 8 |
Hb_001789_190 |
0.0920226784 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 9 |
Hb_023480_010 |
0.0931333065 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_001500_050 |
0.0932645841 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 11 |
Hb_003878_110 |
0.0954734426 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 12 |
Hb_008695_020 |
0.0956424459 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 13 |
Hb_000044_040 |
0.0961408447 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 14 |
Hb_000189_330 |
0.0961424955 |
- |
- |
PREDICTED: ER membrane protein complex subunit 2 [Jatropha curcas] |
| 15 |
Hb_004097_070 |
0.0973316728 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 16 |
Hb_000260_100 |
0.100140683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000498_010 |
0.1010988038 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_001001_080 |
0.1013075312 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 19 |
Hb_002997_080 |
0.1017378184 |
- |
- |
PREDICTED: uncharacterized protein LOC105631115 [Jatropha curcas] |
| 20 |
Hb_002684_040 |
0.1018478345 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |