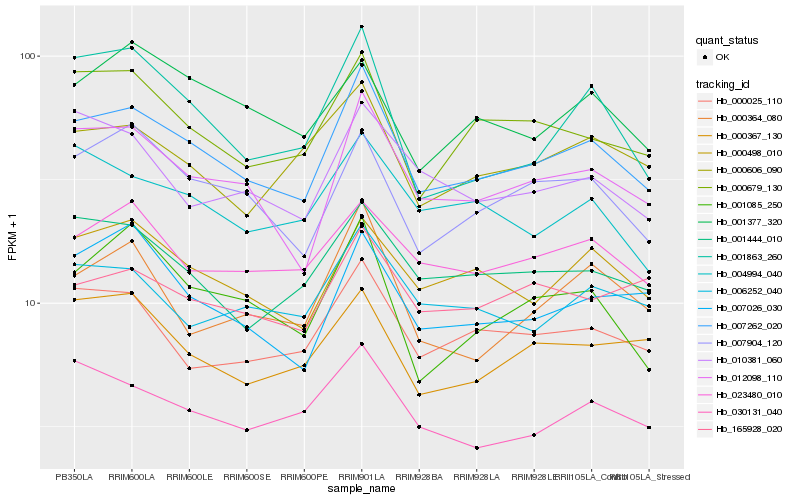
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007262_020 |
0.0 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 2 |
Hb_012098_110 |
0.0584371996 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 3 |
Hb_007904_120 |
0.073530133 |
- |
- |
PREDICTED: ALBINO3-like protein 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000498_010 |
0.0745041422 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 5 |
Hb_000679_130 |
0.0784708313 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 6 |
Hb_000025_110 |
0.0802702748 |
- |
- |
hypothetical protein JCGZ_22722 [Jatropha curcas] |
| 7 |
Hb_001444_010 |
0.08068533 |
transcription factor |
TF Family: DDT |
PREDICTED: DDT domain-containing protein DDB_G0282237 isoform X1 [Jatropha curcas] |
| 8 |
Hb_030131_040 |
0.0818852034 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 9 |
Hb_023480_010 |
0.0828569413 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_006252_040 |
0.0853119185 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000606_090 |
0.0860504888 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 12 |
Hb_007026_030 |
0.0861346192 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 13 |
Hb_001085_250 |
0.0866589282 |
transcription factor |
TF Family: G2-like |
PREDICTED: myb family transcription factor APL isoform X2 [Jatropha curcas] |
| 14 |
Hb_004994_040 |
0.0875776125 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 15 |
Hb_000364_080 |
0.0889105466 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000367_130 |
0.0889265051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_010381_060 |
0.0901576075 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 18 |
Hb_001863_260 |
0.0904175212 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_165928_020 |
0.0914398472 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001377_320 |
0.09272395 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH13-like [Jatropha curcas] |