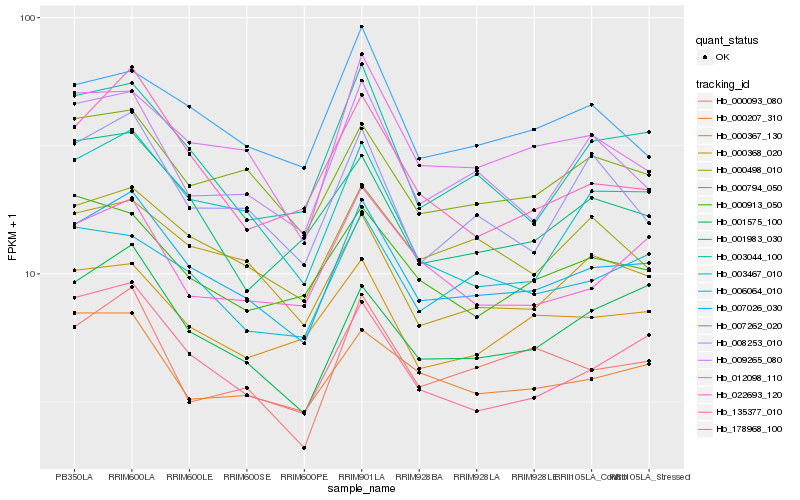
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007026_030 |
0.0 |
- |
- |
rubisco subunit binding-protein alpha subunit, ruba, putative [Ricinus communis] |
| 2 |
Hb_000794_050 |
0.0670196779 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 3 |
Hb_178968_100 |
0.0695034813 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 4 |
Hb_003044_100 |
0.0739144396 |
- |
- |
hypothetical protein F383_08446 [Gossypium arboreum] |
| 5 |
Hb_001575_100 |
0.0742337644 |
- |
- |
PREDICTED: NADPH:adrenodoxin oxidoreductase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000368_020 |
0.0745694905 |
- |
- |
uroporphyrin-III methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_012098_110 |
0.0749911054 |
- |
- |
Ribonuclease 3 [Gossypium arboreum] |
| 8 |
Hb_006064_010 |
0.0768580729 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 4 [Jatropha curcas] |
| 9 |
Hb_000367_130 |
0.0775919274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000207_310 |
0.0782919754 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 11 |
Hb_000498_010 |
0.0787091511 |
- |
- |
PREDICTED: protease Do-like 10, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_009265_080 |
0.0792517813 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 13 |
Hb_003467_010 |
0.0799186708 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 14 |
Hb_135377_010 |
0.0809750073 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62930, chloroplastic-like, partial [Jatropha curcas] |
| 15 |
Hb_000093_080 |
0.0836966636 |
- |
- |
hypothetical protein POPTR_0008s09730g [Populus trichocarpa] |
| 16 |
Hb_022693_120 |
0.0840436017 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 17 |
Hb_008253_010 |
0.084509271 |
- |
- |
PREDICTED: uncharacterized protein LOC105633240 [Jatropha curcas] |
| 18 |
Hb_000913_050 |
0.0855517764 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 19 |
Hb_007262_020 |
0.0861346192 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 20 |
Hb_001983_030 |
0.0865433501 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 12 isoform X2 [Jatropha curcas] |