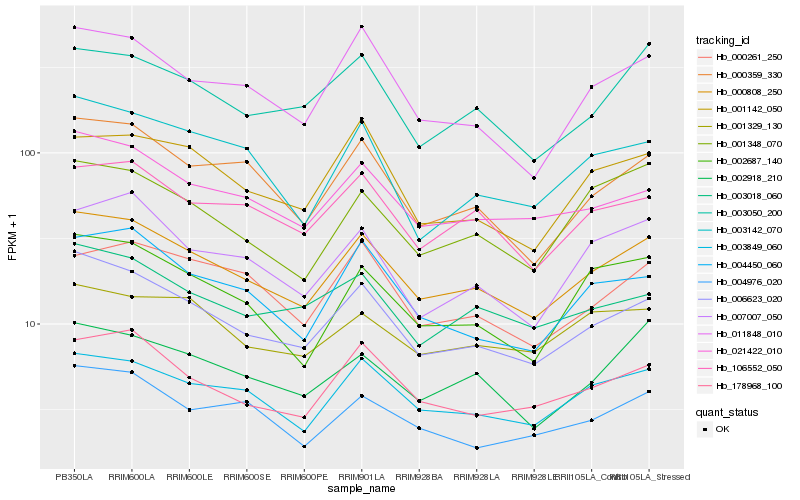
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000808_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647419 [Jatropha curcas] |
| 2 |
Hb_006623_020 |
0.0443746271 |
- |
- |
PREDICTED: protein LURP-one-related 7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003849_060 |
0.0555062961 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 4 |
Hb_021422_010 |
0.0684656565 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000359_330 |
0.0706652603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_178968_100 |
0.0709329254 |
- |
- |
PREDICTED: putative glucose-6-phosphate 1-epimerase isoform X1 [Populus euphratica] |
| 7 |
Hb_106552_050 |
0.0715599486 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 8 |
Hb_011848_010 |
0.0750451086 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 9 |
Hb_002687_140 |
0.076075758 |
- |
- |
hypothetical protein JCGZ_24463 [Jatropha curcas] |
| 10 |
Hb_007007_050 |
0.0786660138 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Malus domestica] |
| 11 |
Hb_003050_200 |
0.0789997605 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 12 |
Hb_004450_060 |
0.0791224014 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002918_210 |
0.0800118471 |
- |
- |
PREDICTED: uncharacterized protein LOC105138897 isoform X2 [Populus euphratica] |
| 14 |
Hb_003018_060 |
0.0818892477 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 15 |
Hb_001348_070 |
0.0822488531 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_004976_020 |
0.082858012 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 17 |
Hb_001329_130 |
0.0831928369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003142_070 |
0.0834037639 |
- |
- |
hypothetical protein F383_14657 [Gossypium arboreum] |
| 19 |
Hb_000261_250 |
0.0853885544 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 20 |
Hb_001142_050 |
0.0855667993 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 12 [Jatropha curcas] |