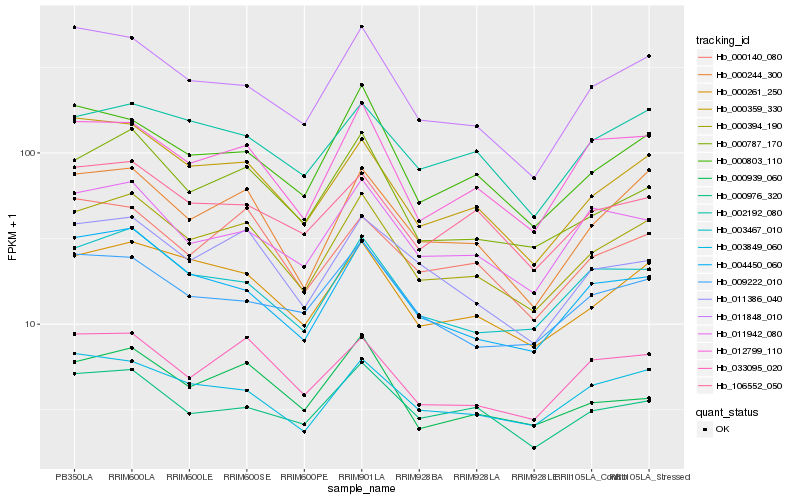
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000394_190 |
0.0 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000261_250 |
0.0547159001 |
- |
- |
elongation factor 1-alpha [Lepidium sativum] |
| 3 |
Hb_000244_300 |
0.0633907211 |
- |
- |
PREDICTED: probable 39S ribosomal protein L17, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000787_170 |
0.0659780136 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 5 |
Hb_003467_010 |
0.0701125221 |
- |
- |
PREDICTED: probable thiol methyltransferase 2 [Jatropha curcas] |
| 6 |
Hb_012799_110 |
0.0715814365 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 7 |
Hb_033095_020 |
0.0731516194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000359_330 |
0.0738470551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011848_010 |
0.0739243495 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 10 |
Hb_003849_060 |
0.0744571979 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 11 |
Hb_002192_080 |
0.076004709 |
- |
- |
hypothetical protein B456_013G069200 [Gossypium raimondii] |
| 12 |
Hb_011386_040 |
0.0767894389 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004450_060 |
0.0799844046 |
- |
- |
PREDICTED: uncharacterized protein LOC105638784 isoform X2 [Jatropha curcas] |
| 14 |
Hb_106552_050 |
0.0810795782 |
- |
- |
PREDICTED: uncharacterized protein LOC105642134 [Jatropha curcas] |
| 15 |
Hb_000803_110 |
0.0812689383 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 16 |
Hb_000140_080 |
0.0820791273 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_011942_080 |
0.0821145245 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 18 |
Hb_000939_060 |
0.0860396039 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 19 |
Hb_009222_010 |
0.0864398531 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_000976_320 |
0.0866026893 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |