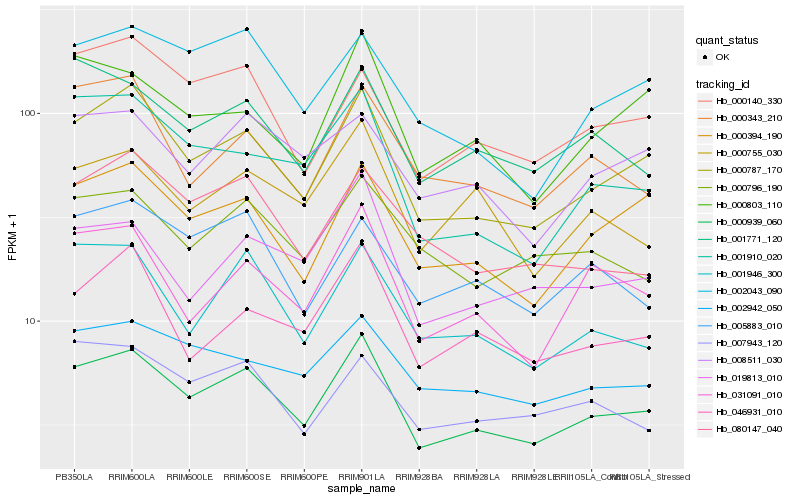
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000939_060 |
0.0 |
- |
- |
PREDICTED: DNA repair protein REV1 [Jatropha curcas] |
| 2 |
Hb_000787_170 |
0.050953653 |
- |
- |
hypothetical protein POPTR_0005s06400g [Populus trichocarpa] |
| 3 |
Hb_000394_190 |
0.0860396039 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_031091_010 |
0.0901042061 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 5 |
Hb_001946_300 |
0.0916618699 |
- |
- |
PREDICTED: F-box protein At-B [Jatropha curcas] |
| 6 |
Hb_001771_120 |
0.0924605426 |
- |
- |
K+ transport growth defect-like protein [Hevea brasiliensis] |
| 7 |
Hb_000140_330 |
0.0938848102 |
- |
- |
PREDICTED: uncharacterized protein LOC105642909 [Jatropha curcas] |
| 8 |
Hb_002942_050 |
0.0939826786 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 9 |
Hb_019813_010 |
0.094846986 |
- |
- |
PREDICTED: uncharacterized protein LOC105646947 [Jatropha curcas] |
| 10 |
Hb_001910_020 |
0.0948889595 |
- |
- |
PREDICTED: ubiquitin-associated domain-containing protein 2 [Jatropha curcas] |
| 11 |
Hb_000755_030 |
0.0953103136 |
- |
- |
PREDICTED: uncharacterized protein LOC105640834 [Jatropha curcas] |
| 12 |
Hb_046931_010 |
0.0978873117 |
- |
- |
hypothetical protein JCGZ_26924 [Jatropha curcas] |
| 13 |
Hb_005883_010 |
0.09816009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007943_120 |
0.0988764517 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 5 isoform X4 [Jatropha curcas] |
| 15 |
Hb_000803_110 |
0.098990094 |
- |
- |
PREDICTED: RNA-binding protein 8A [Jatropha curcas] |
| 16 |
Hb_002043_090 |
0.0991468783 |
- |
- |
hypothetical protein JCGZ_24811 [Jatropha curcas] |
| 17 |
Hb_000796_190 |
0.0997142577 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 18 |
Hb_080147_040 |
0.1005793199 |
- |
- |
hypothetical protein JCGZ_07266 [Jatropha curcas] |
| 19 |
Hb_000343_210 |
0.1014292773 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 20 |
Hb_008511_030 |
0.1015060696 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |