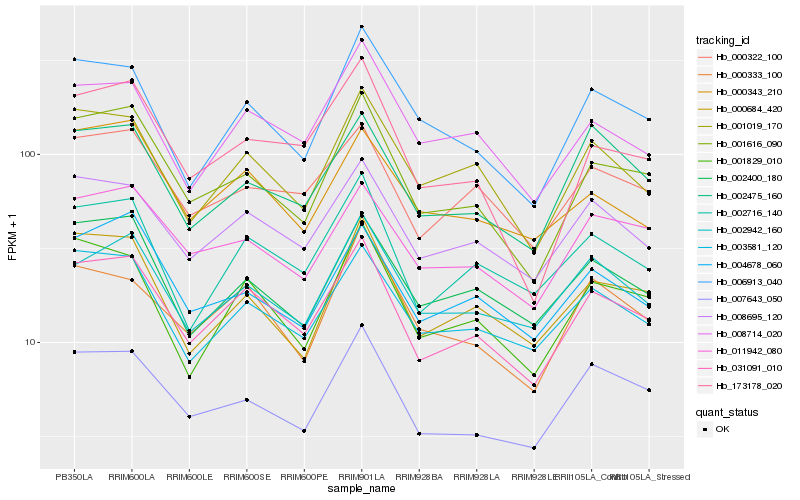
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031091_010 |
0.0 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 2 |
Hb_008695_120 |
0.058455552 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 3 |
Hb_001616_090 |
0.0651454564 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002716_140 |
0.0692867833 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 5 |
Hb_000322_100 |
0.0704152759 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 6 |
Hb_003581_120 |
0.0712618472 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 7 |
Hb_007643_050 |
0.0727108645 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_001829_010 |
0.0743928331 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001019_170 |
0.0753296056 |
- |
- |
PREDICTED: uncharacterized protein LOC105639322 [Jatropha curcas] |
| 10 |
Hb_011942_080 |
0.0794608696 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 11 |
Hb_002400_180 |
0.080767135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008714_020 |
0.0818525374 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 13 |
Hb_002942_160 |
0.0830080331 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 14 |
Hb_000333_100 |
0.083981113 |
- |
- |
PREDICTED: tubulin--tyrosine ligase-like protein 12 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000684_420 |
0.084461999 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_002475_160 |
0.084471061 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 17 |
Hb_173178_020 |
0.0845221507 |
- |
- |
PREDICTED: vesicle transport v-SNARE 12-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_004678_060 |
0.0851560518 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 19 |
Hb_006913_040 |
0.0870137586 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 20 |
Hb_000343_210 |
0.0886754519 |
- |
- |
ran-binding protein, putative [Ricinus communis] |