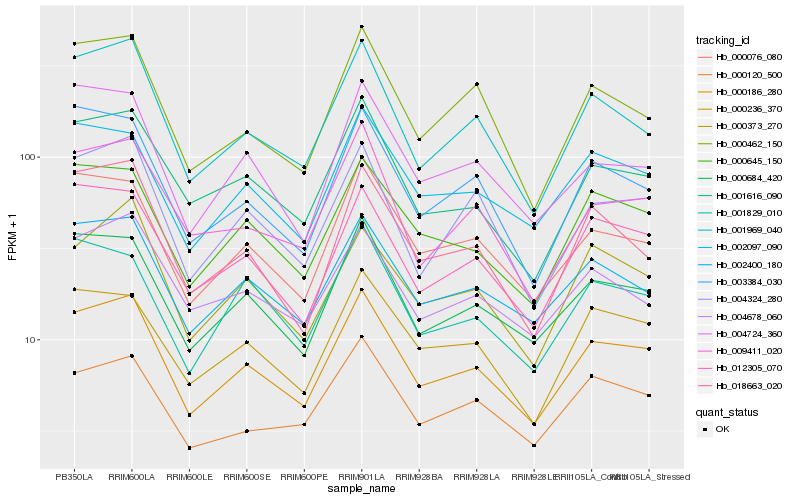
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_280 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000684_420 |
0.052321993 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_001969_040 |
0.060791611 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 4 |
Hb_002400_180 |
0.062014387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000076_080 |
0.0627157417 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 6 |
Hb_000645_150 |
0.06349045 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_012305_070 |
0.0638448287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001616_090 |
0.0648687083 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002097_090 |
0.0669058348 |
- |
- |
Splicing factor 3A subunit, putative [Ricinus communis] |
| 10 |
Hb_000236_370 |
0.0692928237 |
- |
- |
PREDICTED: DNA-directed RNA polymerases I and III subunit RPAC1 [Jatropha curcas] |
| 11 |
Hb_004324_280 |
0.0708365465 |
- |
- |
PREDICTED: plastid division protein PDV1 [Jatropha curcas] |
| 12 |
Hb_000462_150 |
0.0732981721 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Jatropha curcas] |
| 13 |
Hb_009411_020 |
0.0747064937 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit E [Populus euphratica] |
| 14 |
Hb_003384_030 |
0.0760094453 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit beta-like [Jatropha curcas] |
| 15 |
Hb_000120_500 |
0.0771511204 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10-like [Jatropha curcas] |
| 16 |
Hb_000373_270 |
0.0776833477 |
transcription factor |
TF Family: bZIP |
PREDICTED: G-box-binding factor 4 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001829_010 |
0.0791935015 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_004678_060 |
0.0797498787 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 19 |
Hb_018663_020 |
0.0797915433 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004724_360 |
0.0799730516 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |