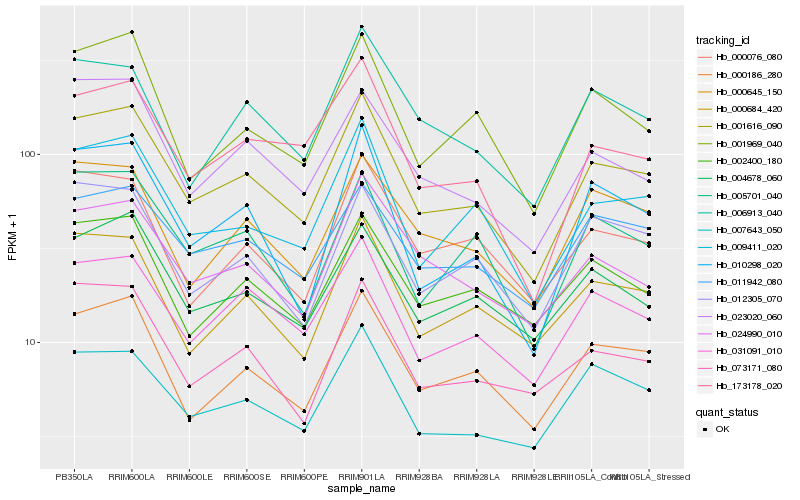
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001616_090 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000186_280 |
0.0648687083 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_009411_020 |
0.0651057424 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit E [Populus euphratica] |
| 4 |
Hb_031091_010 |
0.0651454564 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 5 |
Hb_007643_050 |
0.071079437 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000684_420 |
0.0744186682 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_173178_020 |
0.0775643592 |
- |
- |
PREDICTED: vesicle transport v-SNARE 12-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_004678_060 |
0.0782566668 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 9 |
Hb_001969_040 |
0.0790719959 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 10 |
Hb_073171_080 |
0.0803537004 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_023020_060 |
0.0805341628 |
- |
- |
ubiquitin-conjugating enzyme e2S, putative [Ricinus communis] |
| 12 |
Hb_005701_040 |
0.081321777 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_024990_010 |
0.0818958948 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 14 |
Hb_011942_080 |
0.0820069011 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 15 |
Hb_000645_150 |
0.0820738288 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_012305_070 |
0.0831122969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002400_180 |
0.0840085257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010298_020 |
0.0842630901 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 isoform X4 [Vitis vinifera] |
| 19 |
Hb_000076_080 |
0.085696514 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 20 |
Hb_006913_040 |
0.086358434 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |