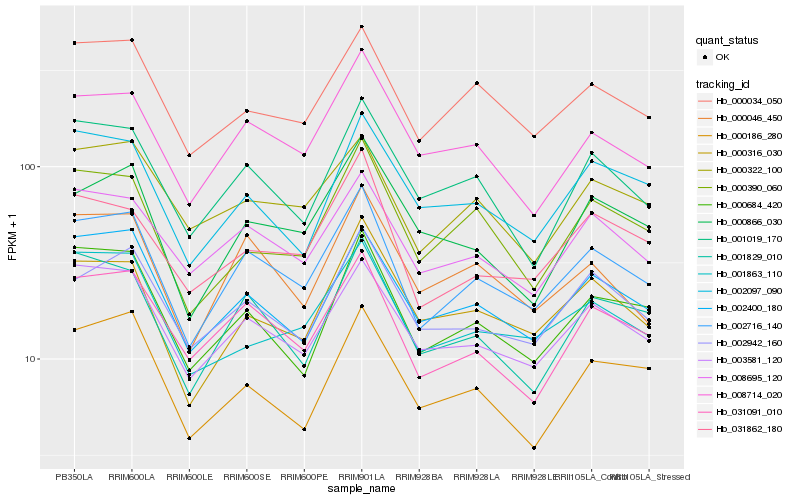
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002716_140 |
0.0 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 2 |
Hb_031091_010 |
0.0692867833 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 3 |
Hb_003581_120 |
0.0694728741 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 4 |
Hb_000034_050 |
0.0741174662 |
- |
- |
PREDICTED: uncharacterized protein LOC105637912 [Jatropha curcas] |
| 5 |
Hb_008714_020 |
0.0753741768 |
- |
- |
PREDICTED: GTP-binding protein SAR1A-like [Jatropha curcas] |
| 6 |
Hb_002400_180 |
0.0754267589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_008695_120 |
0.0754618729 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_001829_010 |
0.0795383706 |
- |
- |
PREDICTED: general transcription factor 3C polypeptide 3-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_000046_450 |
0.0798233209 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 10 |
Hb_002942_160 |
0.0799785221 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 11 |
Hb_000684_420 |
0.0801554894 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000316_030 |
0.0818577795 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_031862_180 |
0.0823456 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 14 |
Hb_001019_170 |
0.0830935267 |
- |
- |
PREDICTED: uncharacterized protein LOC105639322 [Jatropha curcas] |
| 15 |
Hb_000322_100 |
0.0834970503 |
- |
- |
PREDICTED: casein kinase I isoform delta-like [Jatropha curcas] |
| 16 |
Hb_000390_060 |
0.0884293644 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g60570-like [Jatropha curcas] |
| 17 |
Hb_001863_110 |
0.0901716917 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002097_090 |
0.0926954983 |
- |
- |
Splicing factor 3A subunit, putative [Ricinus communis] |
| 19 |
Hb_000866_030 |
0.0930518711 |
- |
- |
hypothetical protein RCOM_1615820 [Ricinus communis] |
| 20 |
Hb_000186_280 |
0.0948898474 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |