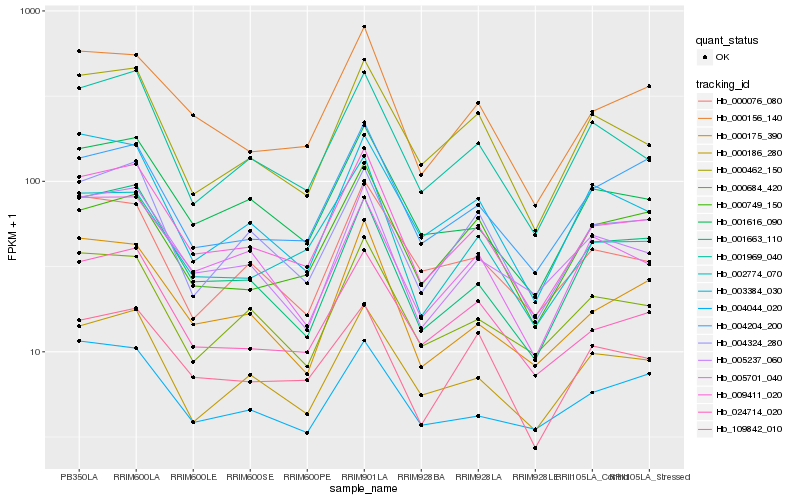
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009411_020 |
0.0 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit E [Populus euphratica] |
| 2 |
Hb_000156_140 |
0.0578280202 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 3 |
Hb_001616_090 |
0.0651057424 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF4 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002774_070 |
0.0673402281 |
- |
- |
hypothetical protein EUGRSUZ_F00307 [Eucalyptus grandis] |
| 5 |
Hb_024714_020 |
0.067619439 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_001969_040 |
0.074563216 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 7 |
Hb_000186_280 |
0.0747064937 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_004324_280 |
0.0789793879 |
- |
- |
PREDICTED: plastid division protein PDV1 [Jatropha curcas] |
| 9 |
Hb_000175_390 |
0.0789989481 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 10 |
Hb_000462_150 |
0.0808676319 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 38 [Jatropha curcas] |
| 11 |
Hb_000684_420 |
0.0829534782 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_005237_060 |
0.0831001694 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 13 |
Hb_004204_200 |
0.0837269233 |
- |
- |
PREDICTED: LYR motif-containing protein 4 [Jatropha curcas] |
| 14 |
Hb_109842_010 |
0.0852863747 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X2 [Jatropha curcas] |
| 15 |
Hb_005701_040 |
0.0889875614 |
- |
- |
cellular nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_003384_030 |
0.0891398155 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit beta-like [Jatropha curcas] |
| 17 |
Hb_000076_080 |
0.0913606752 |
- |
- |
PREDICTED: protein MOS2 [Jatropha curcas] |
| 18 |
Hb_001663_110 |
0.0937802255 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_004044_020 |
0.0939595792 |
- |
- |
PREDICTED: nudix hydrolase 19, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000749_150 |
0.0939830309 |
- |
- |
- |