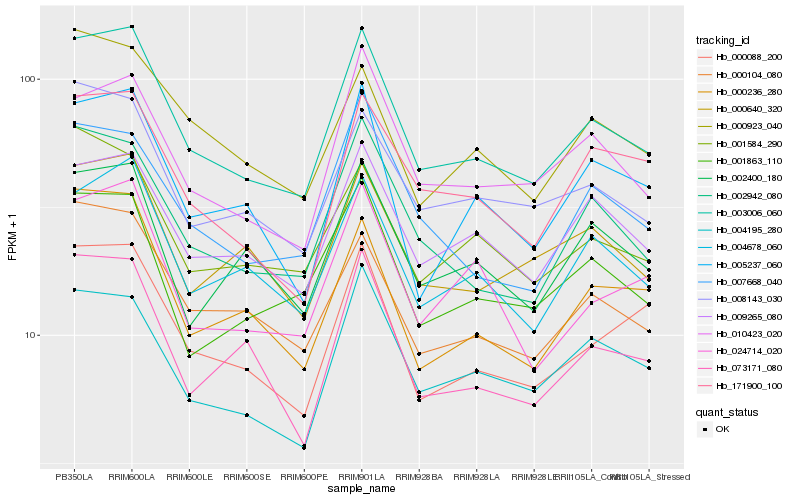
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003006_060 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-splicing factor syf2 [Jatropha curcas] |
| 2 |
Hb_002942_080 |
0.0647251987 |
- |
- |
PREDICTED: signal recognition particle subunit SRP68 [Jatropha curcas] |
| 3 |
Hb_004678_060 |
0.06615964 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 4 |
Hb_000104_080 |
0.0693262832 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 5 |
Hb_010423_020 |
0.0708745788 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC25 homolog [Jatropha curcas] |
| 6 |
Hb_008143_030 |
0.0714199377 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |
| 7 |
Hb_009265_080 |
0.0716542277 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 8 |
Hb_004195_280 |
0.072564608 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 9 |
Hb_000088_200 |
0.0773015833 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 10 |
Hb_000640_320 |
0.0776152988 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007668_040 |
0.0780900067 |
- |
- |
PREDICTED: uncharacterized protein LOC103409082 [Malus domestica] |
| 12 |
Hb_000236_280 |
0.0789452709 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_073171_080 |
0.079278517 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_001584_290 |
0.0801245717 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 15 |
Hb_000923_040 |
0.080679311 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 16 |
Hb_001863_110 |
0.080892929 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002400_180 |
0.0825465562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_171900_100 |
0.0828434177 |
- |
- |
PREDICTED: putative uncharacterized protein DDB_G0270496 [Jatropha curcas] |
| 19 |
Hb_005237_060 |
0.0839336435 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 20 |
Hb_024714_020 |
0.0840918404 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |