| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
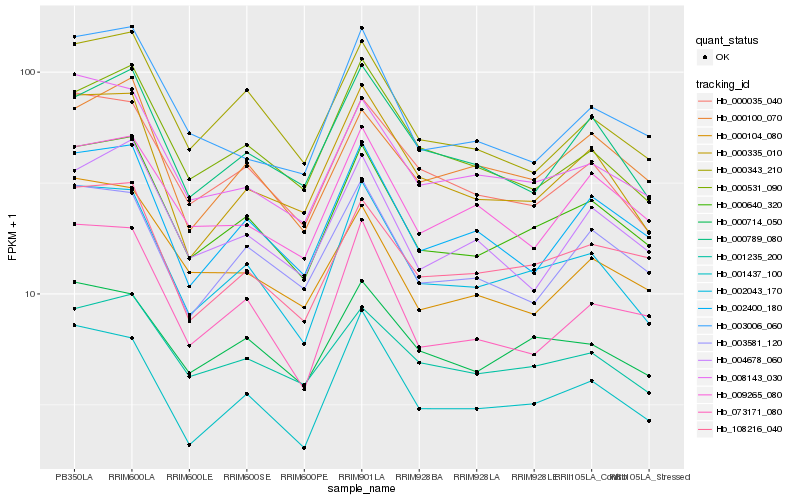
Hb_000640_320 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002043_170 |
0.0559485088 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 3 |
Hb_108216_040 |
0.0595973294 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_008143_030 |
0.0654175317 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000714_050 |
0.0661686503 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 6 |
Hb_002400_180 |
0.0667238011 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000343_210 |
0.0691492024 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000035_040 |
0.0694679953 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 9 |
Hb_001235_200 |
0.0695389408 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003581_120 |
0.0707909721 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 11 |
Hb_000100_070 |
0.0728397576 |
- |
- |
PREDICTED: pre-mRNA-splicing factor ISY1 homolog [Jatropha curcas] |
| 12 |
Hb_004678_060 |
0.0741008733 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 13 |
Hb_001437_100 |
0.0759161553 |
- |
- |
PREDICTED: DNA-binding protein REB1-like [Jatropha curcas] |
| 14 |
Hb_073171_080 |
0.0770256559 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 15 |
Hb_003006_060 |
0.0776152988 |
- |
- |
PREDICTED: pre-mRNA-splicing factor syf2 [Jatropha curcas] |
| 16 |
Hb_000104_080 |
0.0782117921 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 17 |
Hb_000335_010 |
0.078402397 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_009265_080 |
0.0801885295 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 19 |
Hb_000531_090 |
0.0815182724 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 20 |
Hb_000789_080 |
0.0818949508 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |