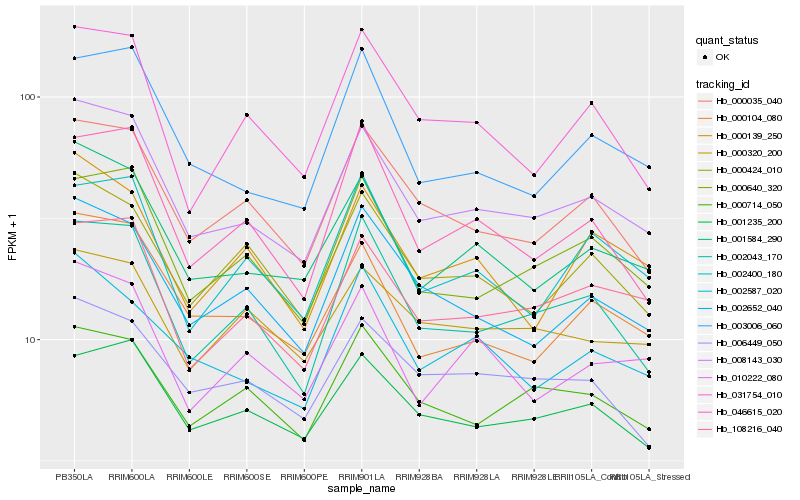
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008143_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001584_290 |
0.0495704674 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 3 |
Hb_108216_040 |
0.0646297558 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 2 [Jatropha curcas] |
| 4 |
Hb_000035_040 |
0.0648837023 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 5 |
Hb_000640_320 |
0.0654175317 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000104_080 |
0.0662052978 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 7 |
Hb_002652_040 |
0.0688963639 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 8 |
Hb_002043_170 |
0.0697557075 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 9 |
Hb_003006_060 |
0.0714199377 |
- |
- |
PREDICTED: pre-mRNA-splicing factor syf2 [Jatropha curcas] |
| 10 |
Hb_000424_010 |
0.0746506833 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002587_020 |
0.076433195 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_000714_050 |
0.0771776761 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 13 |
Hb_001235_200 |
0.0772428396 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006449_050 |
0.077887905 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX2-like [Jatropha curcas] |
| 15 |
Hb_010222_080 |
0.0794710202 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 16 |
Hb_046615_020 |
0.0814265494 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 17 |
Hb_000320_200 |
0.0829264984 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 18 |
Hb_031754_010 |
0.0832350596 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 19 |
Hb_000139_250 |
0.0832376192 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 20 |
Hb_002400_180 |
0.0834569766 |
- |
- |
conserved hypothetical protein [Ricinus communis] |