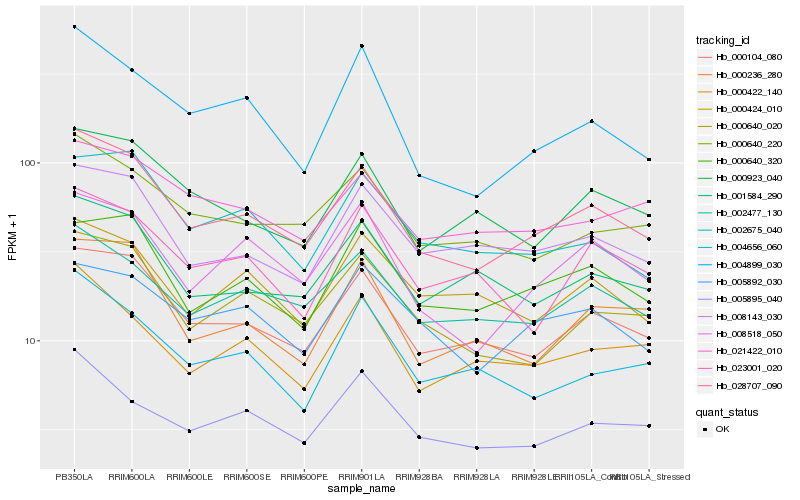
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028707_090 |
0.0 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 2 |
Hb_000104_080 |
0.0588133601 |
- |
- |
PREDICTED: WD repeat domain-containing protein 83 [Jatropha curcas] |
| 3 |
Hb_000640_020 |
0.0785665364 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 4 |
Hb_002477_130 |
0.0801024349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000640_220 |
0.0835857483 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-4 [Jatropha curcas] |
| 6 |
Hb_008518_050 |
0.0841836542 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 7 |
Hb_008143_030 |
0.0842584146 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |
| 8 |
Hb_004899_030 |
0.0874364613 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000640_320 |
0.0882622519 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000236_280 |
0.0889608709 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 22 homolog isoform X1 [Jatropha curcas] |
| 11 |
Hb_001584_290 |
0.0894236963 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 12 |
Hb_000923_040 |
0.0899975987 |
- |
- |
PREDICTED: 15 kDa selenoprotein [Jatropha curcas] |
| 13 |
Hb_005895_040 |
0.0914002217 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 14 |
Hb_000422_140 |
0.0946417306 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002675_040 |
0.0947909814 |
- |
- |
ran-binding protein, putative [Ricinus communis] |
| 16 |
Hb_021422_010 |
0.0969968643 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_005892_030 |
0.0988603593 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 18 |
Hb_000424_010 |
0.0997805741 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 19 |
Hb_004656_060 |
0.099915698 |
- |
- |
PREDICTED: uncharacterized protein LOC105634581 [Jatropha curcas] |
| 20 |
Hb_023001_020 |
0.1003380517 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |