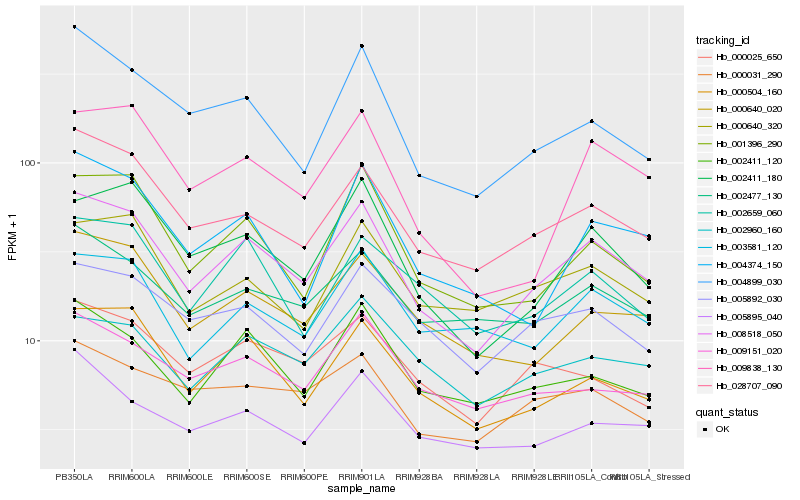
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008518_050 |
0.0 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 2 |
Hb_028707_090 |
0.0841836542 |
- |
- |
PREDICTED: signal peptide peptidase-like 2 [Jatropha curcas] |
| 3 |
Hb_000504_160 |
0.0885810006 |
- |
- |
hypothetical protein POPTR_0001s35340g [Populus trichocarpa] |
| 4 |
Hb_002411_180 |
0.0900542744 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000640_020 |
0.0948111656 |
- |
- |
PREDICTED: probable dimethyladenosine transferase [Jatropha curcas] |
| 6 |
Hb_001396_290 |
0.0967073617 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 7 |
Hb_002411_120 |
0.098745479 |
- |
- |
PREDICTED: uncharacterized protein LOC105631645 [Jatropha curcas] |
| 8 |
Hb_004899_030 |
0.0995077824 |
- |
- |
PREDICTED: 20 kDa chaperonin, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002477_130 |
0.0996977337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000031_290 |
0.1008934136 |
- |
- |
hypothetical protein POPTR_0006s28540g [Populus trichocarpa] |
| 11 |
Hb_005892_030 |
0.1017522346 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 12 |
Hb_009838_130 |
0.1021615183 |
- |
- |
hypothetical protein CISIN_1g010403mg [Citrus sinensis] |
| 13 |
Hb_003581_120 |
0.102261494 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 14 |
Hb_000640_320 |
0.1036628414 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000025_650 |
0.1038210966 |
- |
- |
exosome complex exonuclease rrp45, putative [Ricinus communis] |
| 16 |
Hb_002659_060 |
0.1050933112 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3B [Jatropha curcas] |
| 17 |
Hb_009151_020 |
0.1054027184 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 18 |
Hb_002960_160 |
0.1070016006 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 19 |
Hb_005895_040 |
0.1075599771 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |
| 20 |
Hb_004374_150 |
0.1083596785 |
- |
- |
PREDICTED: uncharacterized protein LOC105650290 isoform X1 [Jatropha curcas] |