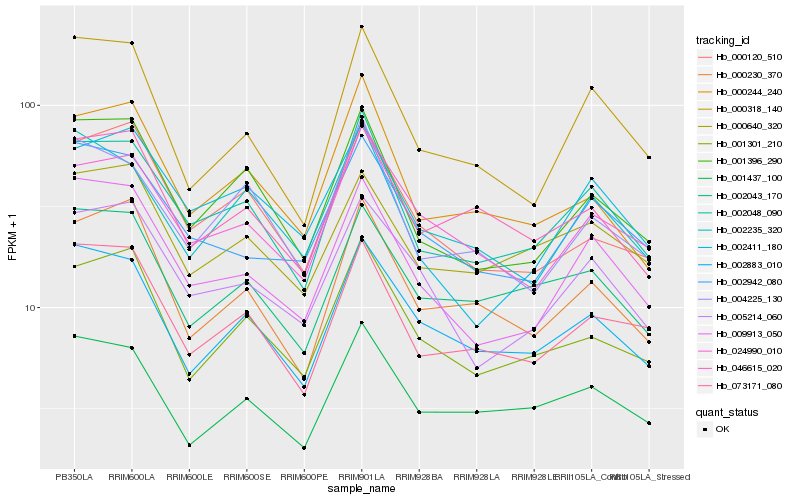
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002048_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 2 |
Hb_001396_290 |
0.0693467358 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 3 |
Hb_073171_080 |
0.0796455125 |
- |
- |
PREDICTED: MMS19 nucleotide excision repair protein homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_009913_050 |
0.0797689719 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 5 |
Hb_000244_240 |
0.0818832938 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_004225_130 |
0.081968232 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002883_010 |
0.0851517243 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 8 |
Hb_024990_010 |
0.0862180818 |
- |
- |
PREDICTED: la protein 1 [Jatropha curcas] |
| 9 |
Hb_001301_210 |
0.0880697072 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 32 [Jatropha curcas] |
| 10 |
Hb_001437_100 |
0.0883072058 |
- |
- |
PREDICTED: DNA-binding protein REB1-like [Jatropha curcas] |
| 11 |
Hb_002043_170 |
0.0891521087 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 12 |
Hb_000318_140 |
0.0900393167 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_000230_370 |
0.0903429209 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 7-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002235_320 |
0.09050797 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000640_320 |
0.0929549256 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002942_080 |
0.093028892 |
- |
- |
PREDICTED: signal recognition particle subunit SRP68 [Jatropha curcas] |
| 17 |
Hb_005214_060 |
0.0936103999 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_046615_020 |
0.09399746 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 19 |
Hb_002411_180 |
0.0956355437 |
- |
- |
PREDICTED: THO complex subunit 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000120_510 |
0.0957603732 |
- |
- |
PREDICTED: RNA polymerase I-specific transcription initiation factor RRN3 isoform X1 [Jatropha curcas] |