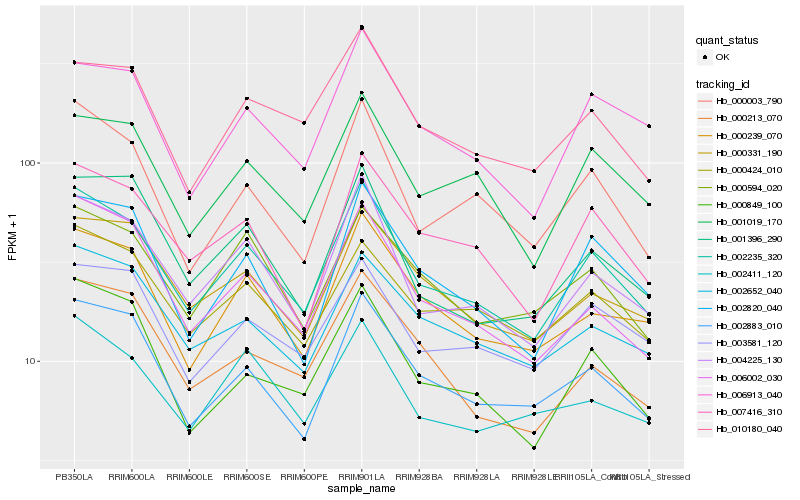
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_320 |
0.0 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004225_130 |
0.0534600747 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105628060 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002883_010 |
0.0693806026 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000239_070 |
0.0725203551 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006002_030 |
0.0736838922 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007416_310 |
0.0737774796 |
- |
- |
PREDICTED: zinc finger protein 622 [Jatropha curcas] |
| 7 |
Hb_001019_170 |
0.0766241834 |
- |
- |
PREDICTED: uncharacterized protein LOC105639322 [Jatropha curcas] |
| 8 |
Hb_000003_790 |
0.0768540984 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 4-like [Jatropha curcas] |
| 9 |
Hb_000331_190 |
0.0769880833 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 10 |
Hb_000424_010 |
0.077374039 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002652_040 |
0.0777485777 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 12 |
Hb_002820_040 |
0.079007312 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000594_020 |
0.0793167328 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 25 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000849_100 |
0.0793195581 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 15 |
Hb_003581_120 |
0.0815368633 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 16 |
Hb_010180_040 |
0.0826440821 |
- |
- |
PREDICTED: prosaposin isoform X1 [Jatropha curcas] |
| 17 |
Hb_006913_040 |
0.0830929489 |
- |
- |
PREDICTED: hydroxyacylglutathione hydrolase cytoplasmic [Jatropha curcas] |
| 18 |
Hb_002411_120 |
0.0835001326 |
- |
- |
PREDICTED: uncharacterized protein LOC105631645 [Jatropha curcas] |
| 19 |
Hb_001396_290 |
0.0838600678 |
- |
- |
hypothetical protein B456_005G166600 [Gossypium raimondii] |
| 20 |
Hb_000213_070 |
0.0859460289 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor At2g41710 isoform X2 [Jatropha curcas] |