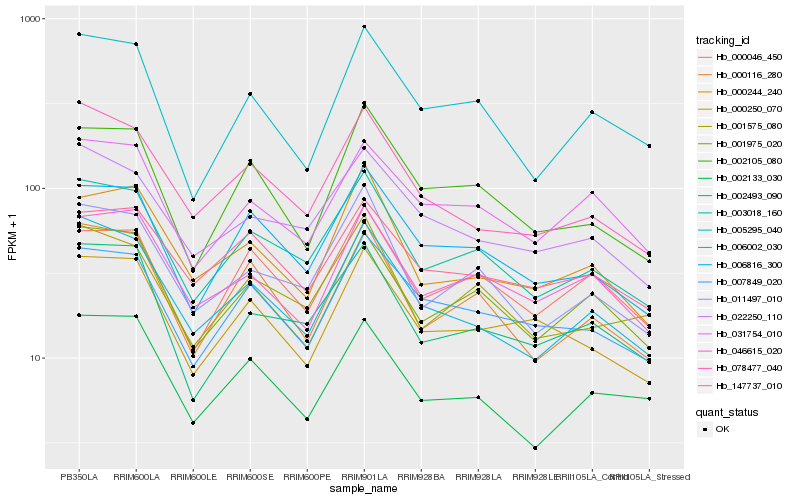
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003018_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628876 [Jatropha curcas] |
| 2 |
Hb_011497_010 |
0.0495519923 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 3 |
Hb_001575_080 |
0.0595560511 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 4 |
Hb_006816_300 |
0.0602852783 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 5 |
Hb_000116_280 |
0.0637160687 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 6 |
Hb_002493_090 |
0.0686227779 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001975_020 |
0.0721703736 |
- |
- |
PREDICTED: uncharacterized protein LOC105637186 [Jatropha curcas] |
| 8 |
Hb_006002_030 |
0.0726928393 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 15 isoform X1 [Jatropha curcas] |
| 9 |
Hb_022250_110 |
0.0727768496 |
- |
- |
PREDICTED: staphylococcal nuclease domain-containing protein 1 [Jatropha curcas] |
| 10 |
Hb_007849_020 |
0.0771658698 |
transcription factor |
TF Family: MYB |
cell division control protein, putative [Ricinus communis] |
| 11 |
Hb_078477_040 |
0.080221491 |
- |
- |
nuclear movement protein nudc, putative [Ricinus communis] |
| 12 |
Hb_000046_450 |
0.0804289638 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 13 |
Hb_002105_080 |
0.0836651902 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 14 |
Hb_031754_010 |
0.0851328789 |
transcription factor |
TF Family: LIM |
zinc ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_046615_020 |
0.0852816315 |
- |
- |
PREDICTED: uncharacterized protein LOC105649409 [Jatropha curcas] |
| 16 |
Hb_005295_040 |
0.0861894648 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP47-like [Jatropha curcas] |
| 17 |
Hb_002133_030 |
0.087699427 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 18 |
Hb_000250_070 |
0.0878903485 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase listerin [Jatropha curcas] |
| 19 |
Hb_147737_010 |
0.0880660596 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000244_240 |
0.0885279011 |
- |
- |
ATP binding protein, putative [Ricinus communis] |