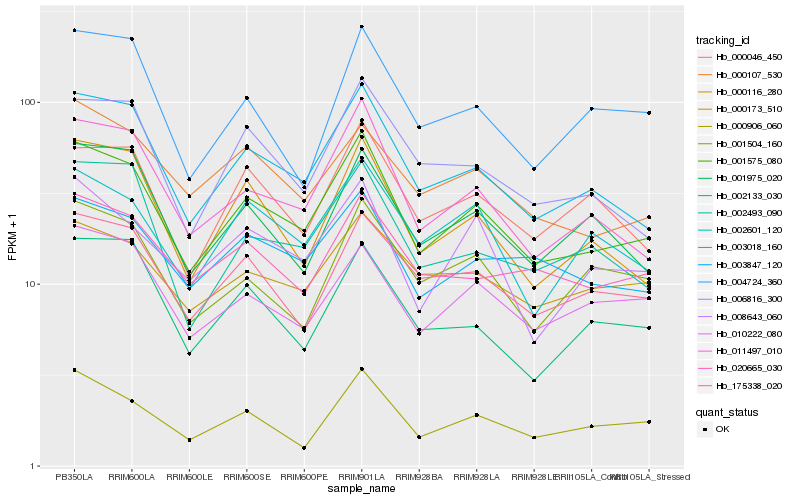
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001575_080 |
0.0 |
- |
- |
PREDICTED: villin-4-like [Jatropha curcas] |
| 2 |
Hb_003018_160 |
0.0595560511 |
- |
- |
PREDICTED: uncharacterized protein LOC105628876 [Jatropha curcas] |
| 3 |
Hb_011497_010 |
0.0718493055 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF126-like [Jatropha curcas] |
| 4 |
Hb_000116_280 |
0.0897807711 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 5 |
Hb_175338_020 |
0.0901735825 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 6 |
Hb_003847_120 |
0.0918045153 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_000906_060 |
0.0936285314 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 8 |
Hb_010222_080 |
0.093937709 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 9 |
Hb_006816_300 |
0.0950967902 |
- |
- |
PREDICTED: importin subunit alpha-2 [Jatropha curcas] |
| 10 |
Hb_020665_030 |
0.0968046775 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002601_120 |
0.0974112196 |
- |
- |
PREDICTED: uncharacterized protein LOC105634921 [Jatropha curcas] |
| 12 |
Hb_000046_450 |
0.097713143 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 13 |
Hb_002493_090 |
0.0981640274 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000173_510 |
0.0992831983 |
- |
- |
PREDICTED: AP-5 complex subunit mu [Jatropha curcas] |
| 15 |
Hb_008643_060 |
0.0993995122 |
- |
- |
unnamed protein product [Coffea canephora] |
| 16 |
Hb_001975_020 |
0.0995990074 |
- |
- |
PREDICTED: uncharacterized protein LOC105637186 [Jatropha curcas] |
| 17 |
Hb_000107_530 |
0.1005931362 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 18 |
Hb_002133_030 |
0.1016437055 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 19 |
Hb_004724_360 |
0.1019107995 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 20 |
Hb_001504_160 |
0.1020148307 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |