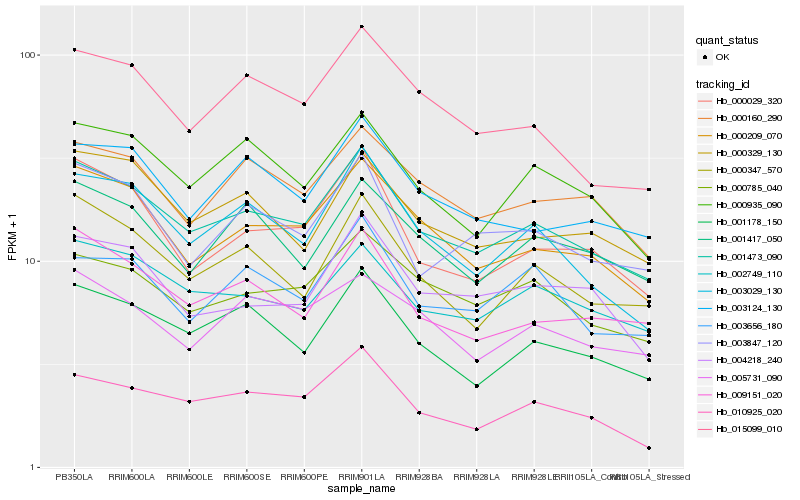
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001473_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 2 |
Hb_000209_070 |
0.0572032193 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 3 |
Hb_002749_110 |
0.0633185611 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001178_150 |
0.0654859264 |
- |
- |
PREDICTED: uncharacterized protein LOC105635484 [Jatropha curcas] |
| 5 |
Hb_000347_570 |
0.0672177483 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12b isoform X1 [Jatropha curcas] |
| 6 |
Hb_009151_020 |
0.0685117612 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 7 |
Hb_000785_040 |
0.0687279097 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 8 |
Hb_000935_090 |
0.068947483 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 9 |
Hb_003029_130 |
0.0703139989 |
- |
- |
carboxypeptidase regulatory region-containingprotein, putative [Ricinus communis] |
| 10 |
Hb_000329_130 |
0.0724127939 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 11 |
Hb_000160_290 |
0.0726001635 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000029_320 |
0.0729479061 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 13 |
Hb_003124_130 |
0.0750910284 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003847_120 |
0.0751941253 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_015099_010 |
0.0754921019 |
- |
- |
PREDICTED: probable glucan 1,3-alpha-glucosidase [Jatropha curcas] |
| 16 |
Hb_010925_020 |
0.0772044437 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 17 |
Hb_001417_050 |
0.0790593864 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 18 |
Hb_004218_240 |
0.0804066593 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 1 [Jatropha curcas] |
| 19 |
Hb_003656_180 |
0.0805391966 |
- |
- |
PREDICTED: uncharacterized protein LOC105637927 [Jatropha curcas] |
| 20 |
Hb_005731_090 |
0.0810072767 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |