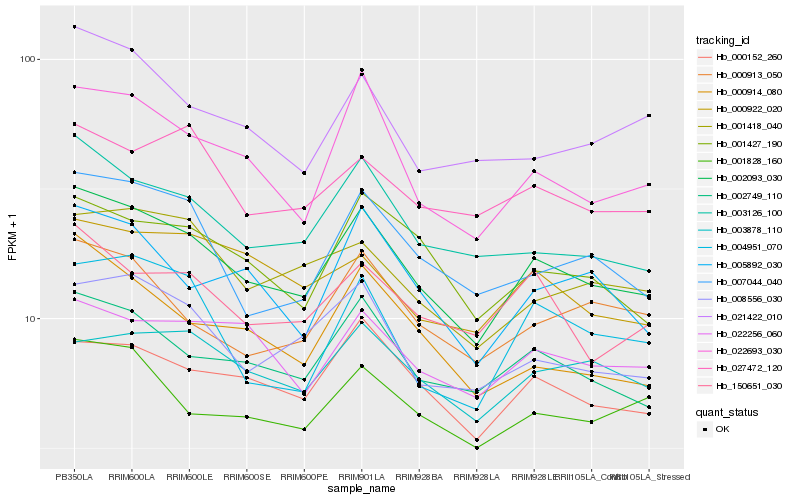
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002093_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 2 |
Hb_002749_110 |
0.0623280943 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003126_100 |
0.0695516194 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 4 |
Hb_022693_030 |
0.069637709 |
- |
- |
PREDICTED: zinc finger protein GIS2 [Jatropha curcas] |
| 5 |
Hb_000152_260 |
0.071608783 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 6 |
Hb_008556_030 |
0.0748542243 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001828_160 |
0.0767691261 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 8 |
Hb_007044_040 |
0.0792851 |
- |
- |
PREDICTED: uncharacterized protein LOC105646814 [Jatropha curcas] |
| 9 |
Hb_001427_190 |
0.0794833228 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 10 |
Hb_005892_030 |
0.0816996042 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 11 |
Hb_000913_050 |
0.0819197265 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 12 |
Hb_022256_060 |
0.0825919135 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 13 |
Hb_150651_030 |
0.0827482186 |
- |
- |
PREDICTED: metallophosphoesterase 1-like [Jatropha curcas] |
| 14 |
Hb_001418_040 |
0.0839494197 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 15 |
Hb_000914_080 |
0.0859158374 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_004951_070 |
0.0870726951 |
desease resistance |
Gene Name: AAA |
Stage III sporulation protein AA, putative [Ricinus communis] |
| 17 |
Hb_027472_120 |
0.0871138013 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_021422_010 |
0.0875835685 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000922_020 |
0.0883884428 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_003878_110 |
0.0885596135 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |