| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
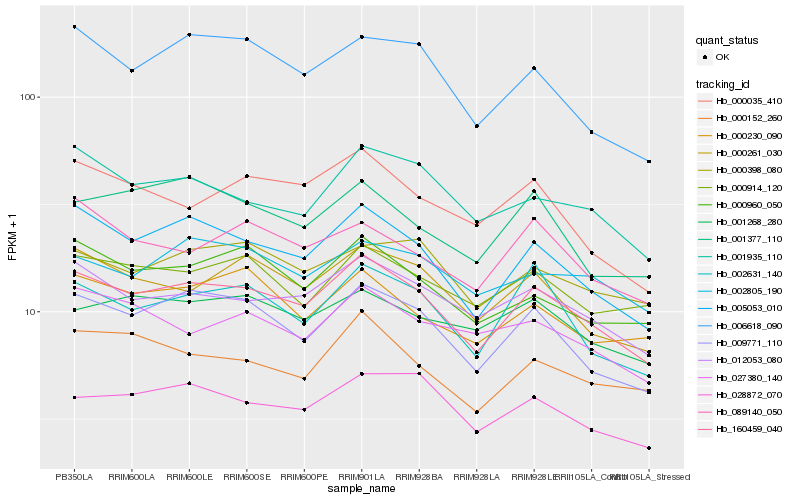
Hb_160459_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 2 |
Hb_005053_010 |
0.0469732987 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 3 |
Hb_009771_110 |
0.0521042291 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 4 |
Hb_012053_080 |
0.0562710269 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 5 |
Hb_000261_030 |
0.0631489256 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 6 |
Hb_002631_140 |
0.0671574414 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 12, chloroplastic [Jatropha curcas] |
| 7 |
Hb_028872_070 |
0.0675147194 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001935_110 |
0.0693624433 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 9 |
Hb_000152_260 |
0.0694906861 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 10 |
Hb_000960_050 |
0.0702400783 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 11 |
Hb_006618_090 |
0.0721674481 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 12 |
Hb_000914_120 |
0.0743004416 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000230_090 |
0.0747677987 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000035_410 |
0.0749362117 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 15 |
Hb_000398_080 |
0.0753147879 |
- |
- |
tip120, putative [Ricinus communis] |
| 16 |
Hb_002805_190 |
0.0770861123 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 17 |
Hb_089140_050 |
0.0773121727 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 18 |
Hb_001377_110 |
0.0774315475 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 19 |
Hb_001268_280 |
0.0782576487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_027380_140 |
0.0783390551 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |