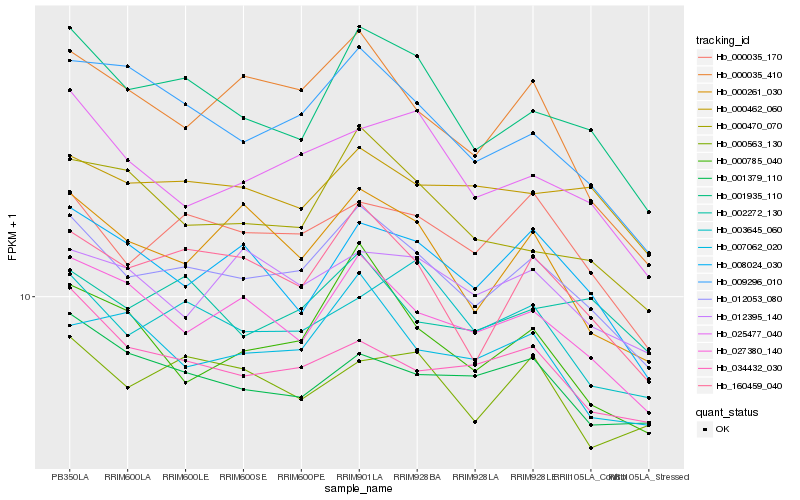
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012053_080 |
0.0 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 2 |
Hb_001935_110 |
0.0546177147 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 3 |
Hb_160459_040 |
0.0562710269 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 4 |
Hb_000035_170 |
0.0570057127 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 5 |
Hb_027380_140 |
0.0597867044 |
- |
- |
PREDICTED: uncharacterized protein LOC105634023 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009296_010 |
0.0672389273 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 7 |
Hb_025477_040 |
0.0695994522 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 8 |
Hb_000261_030 |
0.0699960101 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 9 |
Hb_008024_030 |
0.0707179731 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105648348 [Jatropha curcas] |
| 10 |
Hb_007062_020 |
0.0720054211 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 11 |
Hb_000470_070 |
0.0720869421 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 12 |
Hb_000035_410 |
0.072195696 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 13 |
Hb_000462_060 |
0.0722000024 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 14 |
Hb_034432_030 |
0.0725435947 |
- |
- |
PREDICTED: probable glycosyltransferase At5g20260 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000785_040 |
0.0726642431 |
- |
- |
PREDICTED: uncharacterized protein LOC105628919 [Jatropha curcas] |
| 16 |
Hb_003645_060 |
0.075269313 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002272_130 |
0.0761751723 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 18 |
Hb_012395_140 |
0.076260204 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 19 |
Hb_001379_110 |
0.0763245753 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |
| 20 |
Hb_000563_130 |
0.0766583918 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |