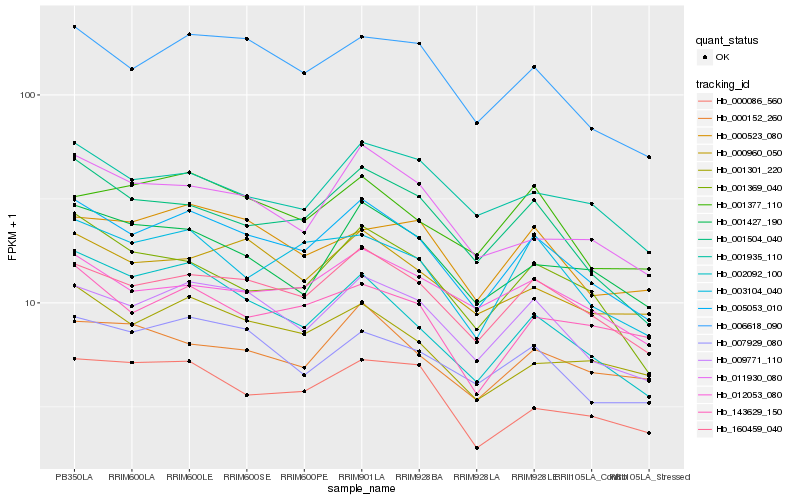
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005053_010 |
0.0 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 2 |
Hb_160459_040 |
0.0469732987 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 3 |
Hb_009771_110 |
0.0480623557 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 2 [Jatropha curcas] |
| 4 |
Hb_006618_090 |
0.0595082856 |
- |
- |
PREDICTED: FAM10 family protein At4g22670 [Jatropha curcas] |
| 5 |
Hb_001504_040 |
0.0709236736 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 6 |
Hb_007929_080 |
0.0727713736 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 7 |
Hb_003104_040 |
0.0734592555 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 8 |
Hb_001369_040 |
0.0764832705 |
- |
- |
PREDICTED: probable isoaspartyl peptidase/L-asparaginase 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000960_050 |
0.0768928016 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 10 |
Hb_001301_220 |
0.0769075888 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 11 |
Hb_001427_190 |
0.0794782372 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 12 |
Hb_002092_100 |
0.0795145886 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 13 |
Hb_001377_110 |
0.0801054393 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 31-like [Jatropha curcas] |
| 14 |
Hb_011930_080 |
0.0812726928 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 15 |
Hb_001935_110 |
0.081367074 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 16 |
Hb_000086_560 |
0.0829861247 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000152_260 |
0.0831762351 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 18 |
Hb_012053_080 |
0.0838227022 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 19 |
Hb_143629_150 |
0.0839471625 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 20 |
Hb_000523_080 |
0.0843486801 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |