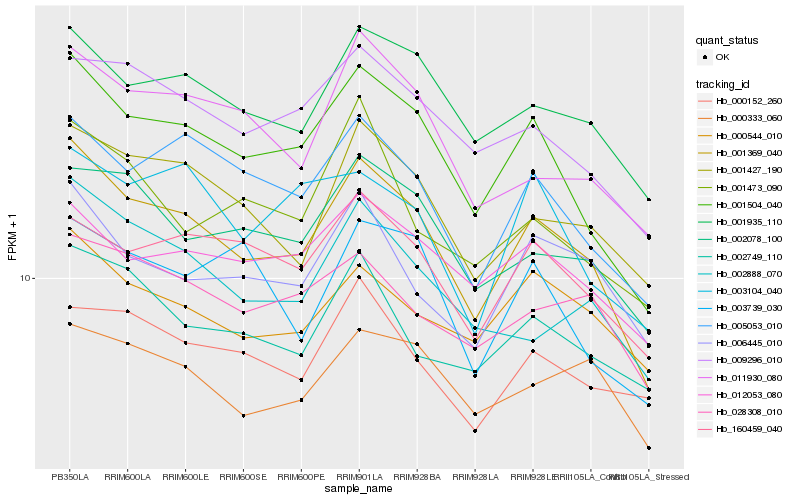
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_040 |
0.0 |
- |
- |
PREDICTED: probable isoaspartyl peptidase/L-asparaginase 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001504_040 |
0.0539802422 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 3 |
Hb_001935_110 |
0.0754081828 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 4 |
Hb_005053_010 |
0.0764832705 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 5 |
Hb_001427_190 |
0.078007415 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 6 |
Hb_002078_100 |
0.0826335598 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 7 |
Hb_009296_010 |
0.082883858 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor GTP-binding subunit ERF3A isoform X3 [Jatropha curcas] |
| 8 |
Hb_006445_010 |
0.0877706064 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 9 |
Hb_002888_070 |
0.0879943554 |
- |
- |
PREDICTED: tubulin-folding cofactor E isoform X1 [Pyrus x bretschneideri] |
| 10 |
Hb_160459_040 |
0.0880767306 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 11 |
Hb_011930_080 |
0.0894411367 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 12 |
Hb_028308_010 |
0.090508437 |
- |
- |
PREDICTED: probable protein S-acyltransferase 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002749_110 |
0.0915192323 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000544_010 |
0.0930872902 |
- |
- |
PREDICTED: GPI mannosyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_012053_080 |
0.0932545708 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 16 |
Hb_000333_060 |
0.0941079737 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 17 |
Hb_003104_040 |
0.0975441127 |
- |
- |
PREDICTED: insulin-degrading enzyme isoform X1 [Jatropha curcas] |
| 18 |
Hb_003739_030 |
0.0993550731 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 19 |
Hb_001473_090 |
0.0997334884 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 20 |
Hb_000152_260 |
0.1002181109 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |